Translate this page into:
Molecular Docking Insights of Newly Synthesized Schiff Base Monomers and Evaluating the Anticancer Activity of Their Polymers
Address for correspondence A. Mohammed Ibrahim, PhD, PG & Research Department of Chemistry, The New College, Affiliated to the University of Madras, Chennai, India (e-mail: mdibrahimchemist@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
The molecular docking technique has shown efficacy with small molecules but faces challenges when applied to macromolecules. To overcome this limitation, a focused approach targeting the active repeat units (monomers) of macromolecules was adopted. This study synthesized ten new dihydroxy Schiff base monomers (SBM1-SBM10) featuring azo moieties and alkoxy side groups. These were attached to human 3-alpha-hydroxysteroid dehydrogenase type 3 (4XO6), a protein linked to breast cancer, using molecular docking via the AutoDock tool.
Materials and Methods
The synthesis of dihydroxy Schiff base monomers SBM1-SBM10 with azo moieties and alkoxy side groups was carried out. These synthesized monomers were then docked with human 3-alpha-hydroxysteroid dehydrogenase type 3 (4XO6) utilizing AutoDock. Among these, the most promisingly docked monomer, SBM8, was selected for further experimentation. SBM8 was polymerized with terephthaloyl chloride to produce a novel polyester termed PolySyringaldehydeDiaminodiphenylSulfone (PSDS). The anticancer activity of PSDS was assessed using the MCF7 human breast cancer cell line. Concurrently, its cytotoxicity was evaluated via the MTT assay employing a normal VERO cell line.
Results
The molecular docking analysis revealed the best-docked monomer, SBM8, which was subsequently used for the synthesis of PSDS. The newly formed polyester, PSDS, demonstrated significant anticancer properties against the MCF7 human breast cancer cell line. Simultaneously, the cytotoxicity evaluation on the normal VERO cell line indicated a favorable safety profile for PSDS.
Conclusion
The study's findings highlight the successful synthesis and docking of dihydroxy Schiff base monomers with 4XO6, resulting in the creation of PSDS. This newly synthesized polyester, PSDS, exhibited promising anticancer activity against the MCF7 cell line while demonstrating minimal cytotoxicity towards normal VERO cells. These results suggest the potential of PSDS as a targeted therapeutic agent against breast cancer, warranting further investigation and development.
Keywords
docking
monomers
anticancer activity
polymers
breast cancer
Introduction
Schiff bases are compounds that contain an azomethine (–C = N–) linkage formed through the condensation of an aldehyde and an amine. They have various applications in fields such as pigments, dyes, catalysts, intermediates in organic synthesis, chemical sensors, and pharmacy. Schiff base aromatic polyester is also reported to exhibit good biological activities. Therefore, polyazomethine esters have been evaluated as anti-inflammatory, antioxidant, and antitumor agents through molecular docking studies.1,2,3,4,5
Breast cancer is the most common malignancy in women, and MCF7 cell lines are commonly used for in vitro analysis.6 Molecular docking analysis can be employed to model the interaction between a small molecule and a protein at the atomic level, enabling us to characterize the behavior of small molecules at the binding site of target proteins and elucidate fundamental biochemical processes.7
Molecular docking involves generating multiple possible conformations of the ligand within the protein binding site. Therefore, the availability of the three-dimensional structure of the molecular target is a necessary condition. This structure can be experimentally solved (e.g., by X-ray crystallography or nuclear magnetic resonance [NMR]) or obtained through computational techniques (e.g., homology modeling).8 Accurate prediction of ligand-target complexes is crucial in modern structure-based drug design.9 Additionally, ligand matching and corresponding scoring can confirm the binding efficiency of each molecule at the binding site.10 The docking process comprises two basic steps: (1) predicting the ligand's conformation, position, and orientation within the binding site (commonly referred to as a pose) and (2) assessing the binding affinity. These steps involve sampling methods and evaluation schemes.11
In the present study, 10 synthetic compounds were synthesized using previously reported methods and evaluated for anticancer activity using human 3-alpha-hydroxysteroid dehydrogenase type 3. The best-docked monomer, SBM8, was polymerized into polyester (PSDS) using terephthaloyl chloride, and its anticancer activity was evaluated using the MTT assay.
The 10 monomers, SBM1-SBM10 (►Fig. 1), were synthesized using the previously described method12 and docked to human 3-alpha-hydroxysteroid dehydrogenase type 3 using the AutoDock tool. The Lamarckian genetic algorithm was employed, utilizing the conformational changes of molecules after in situ optimizations. A quasi-blind docking method was used to define the potential inhibitor binding sites on the surface of human 3-alpha-hydroxysteroid dehydrogenase type 3. It has been demonstrated that the B-chain of human 3-alpha-hydroxysteroid dehydrogenase type 3 interacts with 5-alpha-dihydrotestosterone (5-alpha-DHT) and reduces MCF7 cell growth.13

- Structure of two new series of Schiff base monomers (SBM1-SBM10).
Therefore, the target protein, human 3-alpha-hydroxysteroid dehydrogenase type 3, was used in molecular docking analysis to search for potential inhibitors with the lowest possible binding energy. The crystal structure of human 3-alpha-hydroxysteroid dehydrogenase type 3 (PDB entry 4XO6) was derived and employed for the docking experiments. The target protein was generated by editing the PDB file and removing the heteroatoms (nicotinamide adenine dinucleotide phosphate [NADP]), 1,2-ethanediol, (3beta, 5alpha)-3-hydroxyandrostan-17-one, and all water molecules from the complex structure.
Gasteiger charges were assigned to the structure of human 3-alpha-hydroxysteroid dehydrogenase type 3 using tools from the AutoDock suite. The induced fit docking interface was utilized for docking purposes, and the ligands were treated as flexible. A grid box for ligand docking experiments was centered on the protein molecule, with a spacing of 0.375 and x, y, and z coordinates of 12.077, 13.052, and –30.711, respectively, resulting in a total of 126,126,126 grid points. Additional experimental parameters employed in AutoDock included a population size of 300, the number of generations was set to 27,000, and the number of evaluations was set to 20,000,000. The docking runs were performed 50 times with a mean square deviation cutoff of 1.
Cell Lines and Culture
The MCF-7 human breast cancer cell line was obtained from the King Institute of Preventive Medicine and Research, Chennai, India. The cells were cultured in a culture flask using Minimum Essential Medium supplemented with 3% L-glutamine, 10% fetal bovine serum (FBS), penicillin (100 IU/mL), streptomycin (100 μg/mL), amphotericin B, and 7.5% sodium bicarbonate. The cells were grown in a vented T25 mL culture flask and incubated at 37°C in a 5% CO2 incubator.
After 3 days, the formation of an approximately 80 to 90% confluent monolayer (adherent) was confirmed using a 40× inverted microscope. Subsequently, the cells were subcultured using Trypsin Phosphate Versene Glucose (TPVG) solution in combination with minimal essential medium for further investigations.
Preparation of Stock Solution
Preparation of Stock Solution
The polymer PSDS was dissolved in 1 mL of dimethyl sulfoxide (DMSO) at a concentration of 0.1% (v/v). The volume was then made up to 10 mL using complete medium (MEM) to obtain the extract stock solution at a concentration of 10 mg/mL.
Extract Dilution
The stock solution was further diluted with complete medium to obtain concentrations of 31.25, 62.5, 125, 250, 500, and 1,000 μg/mL. All dilutions were stored in an airtight container until tested.
Cytotoxicity Study: MTT Assay:
The growth inhibition of MCF-7 cells and VERO cells by polyester PSDS was determined using the MTT assay.14 This well-accepted in vitro method is used for screening drugs with cytotoxic activity and helps determine the IC50 concentration of the polymer.
For the assay, cells were harvested and seeded at a density of 105 cells/well in 96-well plates. The plates were then incubated for 72 hours at 37°C with 5% CO2 to allow cell attachment. After 24 hours, different concentrations of the polyester (1, 10, 25, 50, 100 μg/mL), DMSO as vehicle, and a standard 5-fluorouracil (5FU) were added to the cells, followed by incubation for 48 hours. Each concentration was tested in triplicate.
After the incubation period, the medium was replaced with phenol red- and FBS-free medium. Then, 20 μL of MTT dye was added per well, and the plate was wrapped with aluminum foil and incubated for an additional 4 hours. Following incubation, the medium was carefully removed, and 100 μL of DMSO was added to each well to solubilize the formazan crystals. The optical density (OD) was measured at a wavelength of 540 nm.
The percentage of cell inhibition was determined using the following formula:
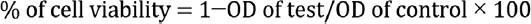
Results and Discussion
The evaluation of the binding affinity of synthetic compounds with human 3-alpha-hydroxysteroid dehydrogenase type 3 was conducted. Among the 10 docked compounds, compound SBM8 exhibited a higher binding affinity, and thus was selected for further studies. The positive control, 5 alpha-DHT (AOX), was used for comparison of binding affinity.
Compound SBM8 was docked with a predicted potential of 24 aromatic carbons and a rotatable bond count of 12. Additionally, SBM10 and SBM6 demonstrated better binding energies of –8.881 and –8.111 kcal/mol, respectively. The binding energy between the ligand–protein complex was –10.94 kcal/mol, with a ligand efficiency of 0.27. The inhibitory potency was determined as 9.56 µM. The intermolecular energy was –14.52 kcal/mol, the van der Waals hydrogen bond dissolution energy was –14.32 kcal/mol, and the electrostatic energy was –0.2 kcal/mol. The total internal energy was –3.25 kcal/mol, while the torsional energy was 3.58 kcal/mol. The unbound or free energy was determined as –3.25 kcal/mol.
Compound SBM8 formed a hydrogen bond between the protein and the ligand molecule at the amino acid LYS270:HZ1, with a bond length of 3.06 Å (►Fig. 2). The positive control AOX-coupled complex formed a hydrogen bond with the amino acid VAL54:OH, with a bond length of 2.04 Å, as reported.

- Interactions from docking performance of the most potent SBM8 with human 3-alpha hydroxysteroid dehydrogenase type 3 (PDB entry 4XO6) visualized in PyMOL.
From the docking studies, it was observed that the SBM8 monomer exhibits superior ligand characteristics compared to the previously studied positive control drug, AOX. SBM8 demonstrates a higher binding affinity and forms stronger bonds with the target macromolecule. The macromolecule plays a crucial role in deactivating the most potent androgen, 5-DHT (AOX control), which subsequently leads to the downregulation of enzyme production and a reduction in MCF7 cell growth.
Previous studies have reported that 5-DHT inhibits proliferation of cancer cell lines, including MCF-7, MDA-MB435S, BT-20, and T47-D. Furthermore, recent research has indicated that 5-DHT can decrease the expression of the efflux transporter, adenosine triphosphate (ATP) binding cassette subfamily G isoform 2 (ABCG2), on the plasma membrane of MCF7 cancer cells, leading to a decline in breast cancer cell growth.15
The SBM8 monomer demonstrates potent inhibitory effects on MCF7 cancer cells and was further tested in vitro using the MTT assay with the MCF7 cell line.
Preparation of polyester PSDS: The polyester PSDS was prepared using a previously reported method12 and the characterization results are the following: yield—72%, FTIR (/cm): 2,915 and 2,846 (–CH2, stretching), 1,734 (C = O-ester, stretching), 1,655 (C = N, extension), 1,612 and 1,547 (C = C-aromatic, extension), and 1,125 (C–O-ester, stretching). 1H NMR (ppm) (DMSO): 8.74 (s, –CH = N), 7.69 (d, 2H), 7.58 (d, 1H), 7.40 (d, 1H), 7.03 (d, 1H), 6.93 (s, 1H), 2.45 (t, 2H), 1.92 (q, 4H), 1.27 (q, 8H), and 3.96 (s, 3H). 13C NMR (ppm) (DMSO): 172.35 (ester carbon); 160.80 (C = N carbon); 149.62, 141.26, 132.12, 125.69, 122.13, 115.62, 111.30, 61.91 (methylene carbon); 56.33 (methoxy carbon); and 42.5, 33.4, 23.17, 23.9, and 21.5 (aliphatic methylene carbons).
Based on the docking results, the SBM8 monomer with the best score was successfully polymerized using terephthaloyl chloride following the described procedure. The cytotoxic activity of the resulting polymer, PSDS, was evaluated against the MCF7 human breast carcinoma cell line and the VERO African green monkey kidney cell line. The IC50 values obtained were 31.2 µg/mL for MCF7 cells and 275 µg/mL for VERO cells (►Figs. 3 and 4). These results indicate that PSDS exhibits cytotoxic effects on cancer cells at relatively low concentrations, while being less toxic to normal cells at higher concentrations, highlighting its potential as an anticancer agent.
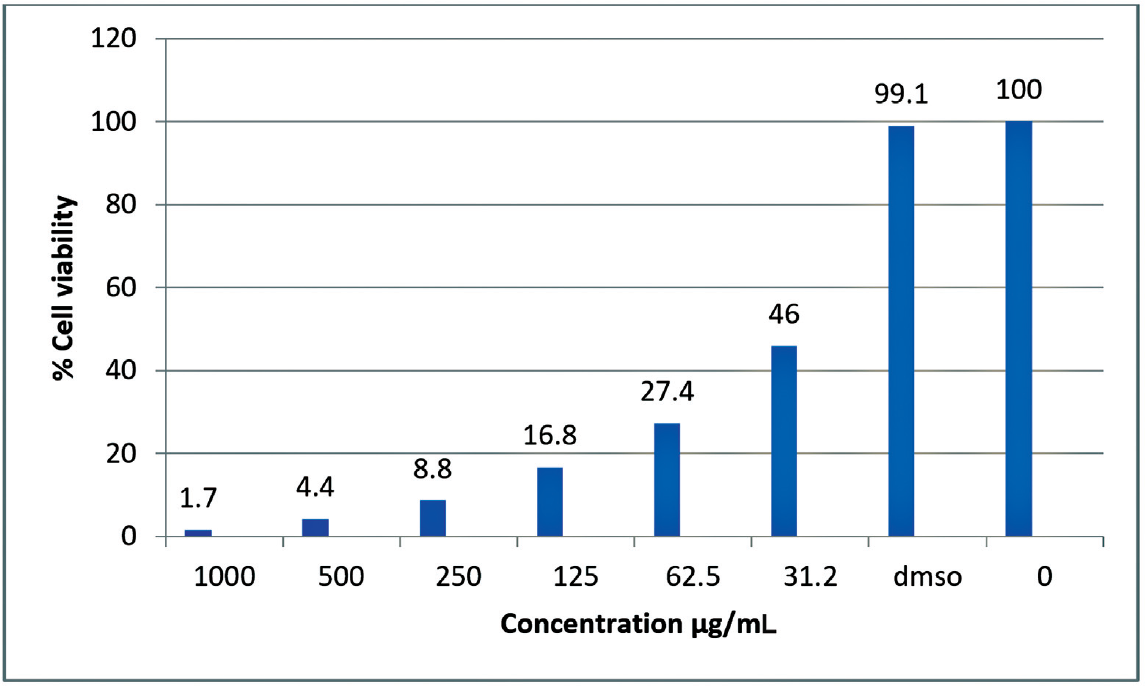
- Graphical representation of percentage of cell viability on the variation of concentration of PSDS.

- Cytotoxic potential of Schiff base PSDS in MCF-7 breast cancer and VERO cell line. (a) Untreated MCF7 control cells, (b) cells treated with 31.2 µg/mL, (c) untreated VERO control cells, and (d) cells treated with 275 µg/mL.
Conclusion
In conclusion, novel Schiff base monomers were synthesized using a previously established method, and docking studies revealed that the SBM8 monomer displayed strong binding affinity with human 3-alpha-hydroxysteroid dehydrogenase type 3. The subsequent in vitro analysis using the MTT assay confirmed the cytotoxic activity of the polymer PSDS derived from SBM8. The in silico docking results align well with the in vitro analysis, demonstrating the potential of using monomers to screen and identify polymers with cytotoxic properties. This research opens up new avenues for screening of polymers based on their monomers for their cytotoxic potential.
Conflict of Interest
None declared.
References
- A review of protein-small molecule docking methods. J Comput Aided Mol Des. 2002;16(03):151-166.
- [CrossRef] [PubMed] [Google Scholar]
- DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15(05):411-428.
- [CrossRef] [PubMed] [Google Scholar]
- Developments in the CHARMM all-atom empirical energy function for biological molecules. Am Chem Soc. 1998;216:U696.
- [Google Scholar]
- AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun. 1995;91(1–3):1-41.
- [CrossRef] [Google Scholar]
- Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225-11236.
- [CrossRef] [Google Scholar]
- The story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res. 2015;35(06):3147-3154.
- [Google Scholar]
- Synthesis and characterization of some new Schiff base polymers. Eur Polym J. 2004;40(04):805-809.
- [CrossRef] [Google Scholar]
- Exploring Protein Flexibility during Docking to Investigate Ligand-Target Recognition [PhD Thesis] 2018
- [Google Scholar]
- A review of protein-small molecule docking methods. J Comput Aided Mol Des. 2002;16(03):151-166.
- [Google Scholar]
- Statistical potentials and scoring functions applied to protein-ligand binding. Curr Opin Struct Biol. 2001;11(02):231-235.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular docking: a powerful approach for structure-based drug discovery. Curr Computeraided Drug Des. 2011;7(02):146-157.
- [CrossRef] [PubMed] [Google Scholar]
- Synthesis and characterization of nanoclay doped PVC/polyester composite film. J Indian Chem Soc. 2019;96:9-13.
- [Google Scholar]
- Human 3α-hydroxysteroid dehydrogenase type 3: structural clues of 5α-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth. Biochem J. 2016;473(08):1037-1046.
- [CrossRef] [PubMed] [Google Scholar]
- Testosterone and 5 α-dihydrotestosterone inhibit in vitro growth of human breast cancer cell lines. Gynecol Endocrinol. 2002;16(02):113-120.
- [CrossRef] [PubMed] [Google Scholar]
- Activity of ABCG2 is regulated by its Expression and localization in DHT and cyclopamine-treated breast cancer cells. J Cell Biochem. 2016;117(10):2249-2259.
- [CrossRef] [PubMed] [Google Scholar]






