Translate this page into:
Breast Cancer in Indian Women: Genetic Risk Factors and Predictive Biomarkers
Address for correspondence Sunita Saxena, MD, DCP, National Institute of Pathology, Indian Council of Medical Research, Safdarjang Hospital Campus, New Delhi, 110029, India (e-mail: sunita_saxena@yahoo.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Breast cancer is the most common cancer among Indian women with a significant increase in incidence in young women. To identify risk factors for breast cancer in young women, study of BRCA1 and BRCA2 germ line mutations was done in a cohort of 204 Indian breast cancer patients. The study showed a total of 18 mutations in 2.94% of the tested patients, 44% BRCA1 and 78% BRCA2 mutations were found unique to the Indian population. Association of low penetrance genes mainly CYP17, VDR gene and AR-CAG repeat polymorphisms with breast cancer risk showed CYP17 A2 and VDR Poly-A L as high risk alleles, the risk of developing breast cancer among women carrying three high-risk alleles is 4.68 (95% confidence interval [CI]: 0.77–28.0; p for trend = 0.10) compared with women carrying none. CYP17 A2 allele was also found associated with development of breast cancer at young age and can also serve as a target for therapy. Betel quid chewing has been found as a significant and independent risk factor for developing breast cancer in North East Indian women which induces genetic alterations leading to breast carcinogenesis. Studies to assess the predictive role of various tumor markers showed that expression of p-glycoprotein in pretreatment biopsy predicts a poor clinical response to neoadjuvant chemotherapy (NACT) in patients having locally advanced breast cancer. The chemotherapy-induced toxicity (vomiting and alopecia) correlated significantly with clinical and immunohistochemical response (reduction in bcl2/bax ratio) and were found to be a cost-effective and reliable predictor of response to NACT. Androgen receptor (AR) has been identified as independent predictive marker for response to NACT in locally advanced breast cancer cases and can serve as novel therapeutic target for triple negative breast cancers.
Keywords
breast cancer
genetic risk factors
predictive biomarkers
Introduction
Breast cancer is currently the most common cancer among Indian women in majority of urban cancer registries, viz. Delhi, Mumbai, Bangalore, Chennai, Ahmadabad, and Thiruvanthapuram (age-adjusted incidence rate [AAR] ranges between 33 and 41 per 100,000 women), and has rapidly overtaken cervical cancer in frequency.1 An analysis of the time trends of breast cancer incidence in India by estimating annual percentage change (APC) in AAR for a 5-year calendar period showed an increase in APC ranging between 0.5 and 2% at different registries (Ahmadabad, Bangalore, Chennai, Mumbai, Nagpur, and Pune). Analysis of APC for age of women by estimating change in age-specific incidence rates (ASR) revealed a 4.24% increase in the younger age group women (15–34 years) compared with 1.6 and 0.8% in age groups of 35 to 44 and 45 to 54 years, respectively, suggesting a significant increase in breast cancer incidence mainly in the younger age group of patients during the last decade.2
These data demonstrate the magnitude of the current health problem associated with breast cancer in the Indian population. Understanding its pathogenesis, morphological features, and epidemiological factors including various risk factors, family history etc., holds a great promise for the treatment, early detection, and prevention of this cancer. A study had been undertaken to describe some of the clinico-pathological features of the breast cancer cases seen at Safdarjung Hospital: a tertiary level hospital in the city of Delhi.3 A detailed analysis of 578 breast cancer cases showed the average age of the patient at presentation to be 47.9 years, 57.9% patients were found below 50 years of age. A positive family history had been found in 22.4% cases, more in patients under 45 years of age. Tumor, node, and metastasis (TNM) staging revealed 78.6% patients presented at locally advanced stage (III B in 36.1%, III A in 27.1%, and II B in16.3%) and metastasis (IV) was found in 7.9% cases.3 Since high percentage of Indian women developed breast cancer at young age (< 50 years) and presented at locally advanced stage, this study had been undertaken to identify the genetic risk factors for detection of breast cancer at early stage and predictive biomarkers to predict response to therapy for proper management.
Genetic Risk Factors for Breast Cancer in Indian Women
Genetic susceptibility to cancer is triggered in several ways; the best understood causal mechanism being due to inactivating germ line mutations in tumor suppressor and DNA repair genes, which lead to accumulation of mutations in oncogenes and cell-cycle checkpoints that are required for uncontrolled cell division. About 5 to 10% of breast and ovarian cancers occur as a result of highly penetrant germ line mutations.
Two major breast cancer susceptibility genes are BRCA1 (MIM 113705, gene bank accession number U14680) and BRCA2 (MIM 600185, gene bank accession number U43746), located on long arms of chromosomes 17 and 13, respectively, and both apparently function as tumor suppressor genes.4,5 Mutations in the BRCA1 and BRCA2 genes were first reported in conjunction with their identification in 1994 and 1996.6-8 Germline mutations in the BRCA1 or BRCA2 tumor-suppressor genes are strong predictors of breast and/or ovarian cancer development. Mutation prevalence varies among different ethnic groups and may be influenced by the potential modifying effects of the patient's own genetic and environmental background.
Prevalence of BRCA Genes Mutations in North Indian Population
A large number of distinct mutations in the BRCA1 and BRCA2 genes have been reported worldwide. To investigate the role of these inherited susceptibility genes in breast cancer risk among Indian women, study of distribution and the nature of BRCA1 and BRCA2 germ line mutations and polymorphisms in a cohort of 204 Indian breast cancer patients and 140 age-matched controls was undertaken at National Institute of Pathology, New Delhi.9
All coding regions and exon–intron boundaries of the BRCA1 and BRCA2 genes were screened by hetero duplex analysis followed by direct sequencing of detected variants.
In total, 18 genetic alterations were identified (►Fig. 1). Three deleterious frame-shift mutations (185delAG in exon 2; 4184del4 and 3596del4 in exon 11) were identified in BRCA1, along with one missense mutation (K1667R), one 5'UTR alteration (22C>G), three intronic variants (IVS10–12delG, IVS13+2T>C, IVS7+38T>C), and one silent substitution (5154C>T). Similarly, three pathogenic protein-truncating mutations (6376insAA in exon 11, 8576insC in exon 19, and 9999delA in exon 27) along with one missense mutation (A2951T), four intronic alterations (IVS2+90T>A, IVS7+75A>T, IVS8+56C>T, and IVS25+58insG), and one silent substitution (1593A>G) were identified in BRCA2 gene. Four previously reported polymorphisms (K1183R, S1613G, and M1652I in BRCA1, and 7470A>G in BRCA2) were detected in both controls and breast cancer patients. Of these, six were pathogenic protein truncating mutations. In addition, several variants of uncertain clinical significance were identified. Among these are two missense variants, one alteration of a consensus splice donor sequence, and a variant that potentially disrupts translational initiation.

- BRCA1 and BRCA2 mutational analysis (A) BRCA1 185 del AG by manual sequencing (B) electropherogram showing BRCA1 22C>G mutation (C) electro pherogram showing BRCA2 8576 insC by automated sequencing.
Of these 18 mutations, six clearly deleterious sequence variants were detected in 2.94% of the tested patients, four of the nine BRCA1 (44%) and seven of the nine BRCA2 mutations (78%) are found unique to the Indian population and are distributed throughout the exons of BRCA1 and BRCA2 genes.
The prevalence of BRCA1/2 mutations in our Indian patient series appeared to be low compared with other Asian countries but is comparatively similar to that reported from Shanghai, China. However, a significant proportion of women who had breast cancer diagnosed at age ≤ 40 years without any family history were also carriers (all variants: 14.2%, known deleterious mutations: 3.8%), which is comparable to other Asian countries. In addition, the present study is in agreement with the findings from a pilot study done on a small independent group of 20 breast cancer patients where 3 out of 5 cases with mutations in BRCA1/2 had early onset disease.10 Thus, it is reasonable to postulate that women with early-onset disease without family history are likely to have a disease associated alteration of the BRCA1 or BRCA2 gene. In the present study, the identified mutations account for a comparatively small proportion of the familial risk of breast/ovarian cancer (all variants: 11.7%, known deleterious mutations: [2.9%]).
Contribution of Low Penetrance Genes to Breast Cancer Susceptibility in Indian Women
Contribution of mutations in highly penetrant genes BRCA1/2 to only 5 to 10% of total breast cancer cases suggests that there are other unidentified genes involved, which may also play role in conferring susceptibility to develop breast cancer at larger scale. Analysis of genetic risk of cancer has shown that most nonhereditary, sporadic cancers develop in genetically predisposed individuals and this predisposition is the result of several low penetrance genes rather than single high penetrant gene mutation. Several classes of low penetrance genes polymorphisms with potential functional significance have been evaluated, including genes involved in (1) carcinogen metabolism (GST family), (2) estrogen metabolite biosynthesis (CYP17), (3) steroid hormone receptor activation (VDR, AR), (4) DNA damage response and replication checkpoint (CHEK2), (5) DNA repair (XRCC1), and (6) alcohol metabolism (ADH3). Because endogenous steroid hormone exposure is known to influence breast cancer risk, genes responsive to such hormones are currently being considered as plausible candidates for low-risk breast cancer genes. Steroid hormones viz. estrogen, progesterone, androgen, and vitamin D are key factors in the development and growth of tumors in hormone dependent tissues specially breast. According to some studies, genetic polymorphisms in genes related to the biosynthesis and metabolism of hormones are associated with an altered risk of breast cancer and other types of cancer. Studies have been undertaken to assess the possible association of low penetrance genes mainly CYP 17, VDR gene, and AR-CAG repeat polymorphisms with breast cancer risk in North Indian women.
CYP17 Gene Polymorphism and Its Association with High-Risk North Indian Breast Cancer Patients
Hormonal risk is one of the nongenetic factors that may contribute to the disease etiology. Several clinical, epidemiological, and experimental studies have proved that estrogen and progesterone play a major role in the growth of normal breast tissue,11 and breast cancer risk is strongly related to exposure to these endogenous steroid hormone levels,12 specially estrogen.13 Because cumulative exposure to circulating estrogen has been linked to increased risk of breast cancer, interest has been focused on the enzymes regulating the biosynthetic and metabolic pathways of these hormones. The estrogen-forming ovary expresses the CYP17 gene,14 which codes for the cytochrome P450c17a enzyme and mediates both steroid activities and functions at key branch points in human steroid genesis.15 A single polymorphism, T > C, in the CYP17 gene creates an additional Sp I- type promoter site (CCACC), which is associated with increased estrogen level.16 The base pair change also creates a recognition site for the MspA1 digestion of a polymerase chain reaction (PCR) product that has been used to designate two alleles, A1 and A2, arbitrarily. It has been reported that women who are heterozygous or homozygous for the CYP 17 A2 allele show comparatively higher serum levels of estradiol than do women with an A1/A1 genotype.17
To investigate the influence of genetic polymorphism of CYP17 on breast cancer susceptibility mainly in high-risk Indian breast cancer patients, targeting early onset breast cancer and familial cases, a total of 242 histopathologically confirmed breast cancer (106 early onset, 80 late onset, and 56 familial) and 212 age-matched controls were investigated for CYP 17 gene polymorphism by PCR-RFLP method at National Institute of Pathology, New Delhi.18 The genotypes identified were assigned as homozygous wild type (A1A1), heterozygous variant (A1A2), and homozygous variant (A2A2; ►Fig. 2). We additionally investigated association of CYP17 polymorphisms with expression of estrogen receptor. The results showed significantly increased (p = 0.001) frequency of heterozygous (A1A2) and homozygous (A2A2) CYP17 genotypes in breast cancer patients (94.3%) than in controls (80.3%). A highly statistically significant increased risk in carriers of homozygous A2 allele was found in young patients (p < 0.001) in comparison with patients having late onset condition (p = 0.260). However, no significant association between the genotype and breast cancer risk was observed among women with strong family history. Further, data had showed that patients (80.6%) with at least one A2 allele exhibit ER-independent cell proliferation, although statistical significance could not be established (p = 0.160).

- (A) MspAI restriction fragments of CYP17 alleles separated by agarose gel electrophoresis. Lane 1: 100 bp ladder, 1 to 12 are samples; Lane 2, 7, and 8: homogygous A1A1 (415bp); Lane 3, 9, 10, and 11: heterozygous A1A2 (415bp, 300bp, 115bp); Lane 1, 6, and 12: homozygus A2A2 (300 bp). (B, C): Electropherogram showing heterozygous (B) and homozygous (C) A2 alleles of CYP17 T>C polymorphism.
VDR Polymorphism(s) and Breast Cancer Risk in North Indians
Identification of 1, 25 (OH)2-D3 and VDR as components of a signaling network that affects breast tissue proliferation, differentiation, and apoptosis raises the possibility that Vitamin D may play a protective role against mammary transformation and common VDR gene variants may be associated with the risk of breast cancer.19-21 This study examined the association between VDR (ApaI/TaqI/Poly-A) genotype breast cancer risk in North Indian population. Genotypes for polymorphism at 3′ end of VDR gene were studied in 160 cases and 140 control subjects using PCR-RFLP method to determine Apa-I and Taq-I polymorphisms and fragment analysis to determine variable length poly-A micro satellite repeats.22
Patient's having long poly-A repeats (χ2 = 87.03, df = 2, p = < 0.001) and homozygous L allele (odds ratio [OR]: 26.97, 95% CI: 12.27–59.17) showed a significant association with disease while no significant association was found with VDR Apa-I (χ2 = 1.00, df2, p = 0.60), and Taq-I polymorphism (χ2 = 0.35, df2, p = 0.83). The results indicate that VDR poly-A genotypic variants confer susceptibility to breast cancer with increasing number of long (L) poly-A allele, while Apa-I polymorphism may modify the individual susceptibility to disease. Analysis of the VDR genotype in relation to covariates has revealed that the LL genotype is significantly associated with high grade breast cancer suggesting that this VDR variant might be associated with disease progression.
The genotype also mediates an increased risk of breast cancer in women with early onset compared with late onset. We also observed significant associations between disease and genotype AATTLL (OR: 5.09; 95% CI: 1.23–20.80; p = 0.02). The decrease in risk in terms of odds ratio from 5.09 to 2.08 even with single (a) in genotype AaTTLL may indicate the protective role of the same in the presence of susceptible (L) allele.
AR-CAG Repeat Polymorphism and Breast Cancer Risk in North Indians
The endogenous steroid hormones (estrogen, progesterone, and androgen) exposure is known to influence breast cancer risk, hence genes responsive to such hormones are currently being considered as plausible candidates for low-risk breast cancer genes. Considerably, little is known about the biological role and clinical significance of androgen and its receptor (AR) expression in breast cancer.23,24 AR protein, functions as a transcription factor (TF) that regulate the transactivation of hormone responsive genes and is thus of specific interest. The exon 1 of AR gene contains trinucleotide repeat polymorphism, CAG (encoding for polyglutamine) which flank the N-terminal domain of the AR protein, where the transactivation activity resides. Remarkably, a CAG trinucleotide repeat is also a target for multiple RNA binding proteins, which have functional impact on AR protein function.6,7 Sparse epidemiologic data suggest that a long AR-CAG repeat yielding a less active AR may be associated with increased risk of breast cancer.25 Polymorphisms in AR-CAG repeat have been intensively studied as determinant of susceptibility to prostate cancer in Indian population26,27; however, its association with breast carcinoma in Indian population is not yet explored.
To determine the role of AR-CAG repeats in risk of breast cancer and their association with response to NACT, genotyping of the AR-CAG repeat region was done in 70 patients and 80 healthy aged matched female controls.28 Genotype carriers with ≥20 CAGn showed decrease of AR mRNA expression, however, no association could be established with the risk (p < 0.47). In patients with locally advanced breast cancer, higher AR mRNA expression was seen prior to NACT in responders (p < 0.02), which decreased significantly after NACT (p < 0.014). Although, expansion of the CAGn in the AR gene did not show any association with breast cancer risk, however, significant correlation was found with response to NACT in locally advanced breast cancer (LABC) patients.
Multigenic Model of Breast Cancer Susceptibility
Considering CYP17 A2 and VDR poly- (A) L as high risk alleles, a multigenic model of breast cancer susceptibility was developed to identify women carrying a combination of alleles, which put them at a relatively higher risk to develop breast cancer. The risk among women carrying three high-risk alleles was 4.68 (95% CI: 0.77–28.0; p for trend = 0.10) compared with those who carried none. The conditional logistic regression analysis revealed that the heterozygous TC genotype for CYP17 and AR1AR2 of AR imparted significantly fourfold risk for the breast cancer risk, in comparison to the referent genotype TT and AR1AR1 (adjusted ORs: 3.705 [1.236–11.106], p < 0.019 and 4.391 [1.324–14.557], p < 0.016, respectively). Gene–gene interaction showed that the combinations TC*AA, TC*Aa, TC*aa, and CC*Aa imparted significantly four- to 15-fold more risk for the breast cancer (4.377 [1.159–16.520], p = 0.029; 4.041 [1.092–14.956], p = 0.036; 15.071 [0.975–232.81], p = 0.052; 4.151 [1.053–16.371], p = 0.042, respectively; ►Table 1). These findings suggest that breast cancer risk has a strong genetic component and support the theory that the underlying mechanism of “complex trait” can be understood using a multigenic model of low penetrant genes. Genes involved in hormone synthesis and metabolizing pathway play significant role in breast cancer development.
| Code (CYP17, VDR, AR) | No. of cases (%) (n = 70) | No. of controls (%) (n = 80) | OR (95% CI) | p-Value | |
|---|---|---|---|---|---|
| No putative high risk genotype | 000 | 2 (2.8) | 3 (3.7) | Ref. | – |
| One putative high risk genotype | 001, 002, 010, 2020, 100, 200 | 13 (18.5) | 27 (33.7) | 0.72 (0.12–4.06) | 1.0 |
| Two putative high risk genotype | 011, 012, 021, 022, 110, 120 201, 202, 210, 220 | 30 (42.8) | 42 (52.5) | 1.07 (0.20–5.69) | 1.0 |
| Three putative high risk genotype | 111, 112, 122, 211, 212, 221, 222 | 25 (37.5) | 8 (10.0) | 4.68 (0.77–28.0) | 0.1 |
Abbreviation: OR, odds ratio.
Note: 0: A1A1, SS, <20<20 CAG repeat; 1: A1A2, SL,<20≥20 CAG repeat; 2: A2A2, LL, ≥20≥20 CAG repeat.
Study of Environmental and Genetic Risk Factors for Breast Cancer in North East India
The several fold difference in incidence of breast cancer between different geographical regions suggest that environmental factors also influence breast cancer risk significantly. Among the identified environmental risk factors in general for cancers, tobacco exposure has been reported as the leading preventable risk factor.29 The Northeast districts of India have the highest incidence of cancers associated with both smoking and smokeless tobacco. The mean age for tobacco use initiation in this region is 18.5 years and the prevalence of tobacco use is estimated as 41% that includes a large number of female chewers too apart from male smokers.30 Epidemiological perspective suggests an increased risk associated with exposure to genotoxic agents during breast development, as the undifferentiated ductal elements of the breast are more susceptible to the action of genotoxins early in life.31 Environmental genotoxic stress like tobacco smoke and smokeless tobacco contain polycyclic aromatic hydrocarbons (PAHs), tobacco-specific nitrosamines, nitrosamino acids, aldehydes, metals, aromatic and heterocyclic amines, and other genotoxic carcinogens.32,33 The concomitant use of betel quid also leads to a 50-fold increase in reactive oxygen species generated.34
The role of tobacco exposure and polymorphisms in detoxification enzymes in breast cancer risk was analyzed using samples collected from patients registered at Dr. B Borooah Cancer Institute, Guwahati. Polymorphisms in five gene variants, GSTT1, GSTM1, GSTP1, TP53 and CYP17, and four environmental exposure variables (tobacco smoking, tobacco chewing, betel quid chewing, and alcohol) were analyzed in 117 breast cancer cases and 174 cancer free controls.35 Multifactor dimensionality reduction (MDR) identified betel quid chewing as the single main risk factor and women with betel quid chewing history had five times the risk of developing breast cancer (OR: 4.78 [2.87–8.00], p = 0.001). In logistic regression analysis, GSTT1 null and GSTM1 null genotypes conferred 41% (0.59 [0.34–1.03] 0.06) and 55% (0.58 [0.30–1.02] 0.05) reduced risk to breast cancer, respectively. However, the risk increased in women with GSTP1 variant G allele, which conferred 1.43 times ([0.96–2.11] 0.07) more risk to breast cancer. MDR analysis revealed, betel quid chewing to be the single factor imparting the main effect (testing accuracy of 0.6851 and cross validation consistency 10/10, p = 0.05). The Figure below depicts the interactions between nine attributes from the MDR analysis via a graphical representation of a “dendrogram” (►Fig. 3). It shows betel quid chewing, GSTT1 and GSTM1 on a separate branch imparting their independent effects to breast cancer risk. The study highlights betel quid chewing as a significant and independent risk factor for developing breast cancer in NE region.
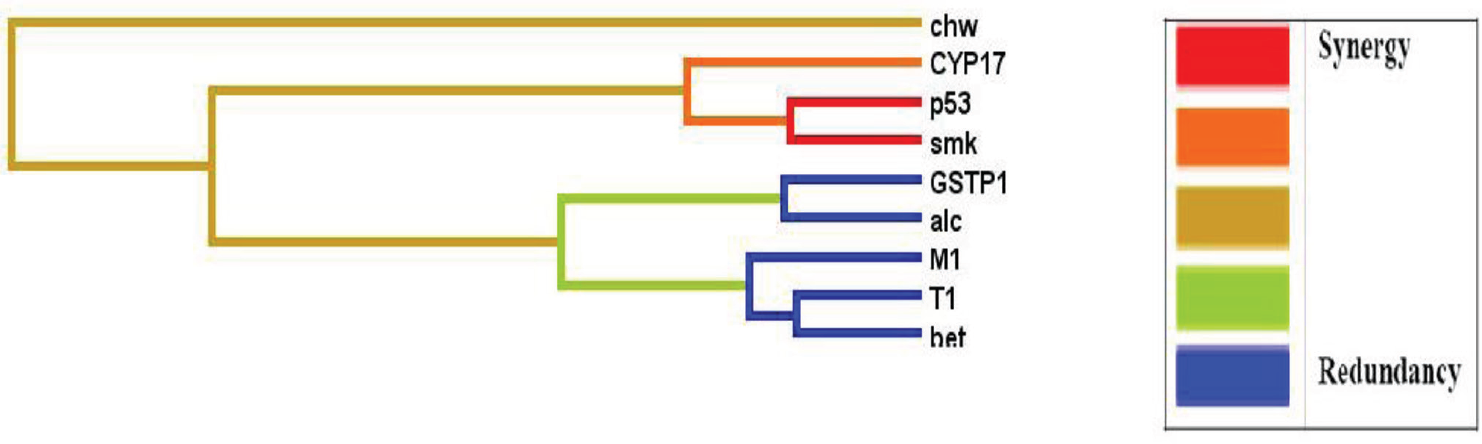
- Interaction dendrogram for the breast cancer dataset: graphical representation of interactions between nine attributes (GST1 (T1), GSTM1 (M1), GSTP1, CYP17, TP53 (p53), tobacco smoking (smk), tobacco chewing (chw), betel quid chewers (bet) and alcohol consumption (alc)) from the multifactor dimensionality reduction analysis using an ‘interaction dendrogram’.
Association of DNA Repair and Cell Cycle Gene Variations with Breast Cancer Risk
Polymorphisms in DNA repair and cell cycle genes contribute to increased breast cancer risk. Their association and interaction in relation to betel quid and tobacco chewing habits needs exhaustive multi-analytical investigation to explain breast cancer predisposition due to DNA damage.
Polymorphism in TP53–72Arg>Pro, RAD51–135G>C, BRCA2, and CCND1-G870A were examined in 204 BC cases and 217 controls from Northeast Indian population.36 Multifaceted analytic approaches were used to explore relationships between polymorphisms, tobacco history, and breast cancer susceptibility. Betel quid chewing was identified as the predominant risk factor. CCND-AA and dominant model showed protection toward breast cancer in BQC (betel-quid chewer) and NBQC (nonbetel-quid chewer). TP53-Pro/Pro genotype showed protection toward breast cancer in NBQC. RAD51-C allele was associated with breast cancer risk in BQC. Two BQC cases had BRCA2 8415G >T:K2729N mutation in exon18.
The hierarchical interaction graph showed (►Fig. 4A), large independent effect of betel quid chewing (9.38%) among environmental factors for total sample set. A strong interaction (1.32%) was seen between RAD51 and TP53. MDR analysis showed best 4 locus model in NBQC. Interaction diagram concurred the interactions between TP53 and RAD51 (1.32%) with independent effect (1.89%) of CCND1 in NBQC (►Fig. 4B). In CART analysis, BQC with CCND1 GG genotype were at risk followed by combination of BQC, CCND1, no smoking, alcohol. Risk was also observed in BQC, CCND1, no smoking, nonalcohol, TP53 combination and BQC, CCND1, no smoking, nonalcohol, TP53 (►Fig. 5). NBQC group showed risk with combination of NBQC and TP53.

- Interaction dendrogram using orange software for the (A) total dataset and (B) the NBQC dataset. tbsmk, tobacco smoking; bqchew, betel-quid chewing.

- Classification and regression tree (CART) analysis for the DNA repair and cell cycle genes and environmental risk factors. Terminal nodes are bordered thick blue. Red bordered odd ratio boxes values are <0.05. BQC, betel-quid chewer; NBQC, non betel-quid chewer; smk, smoking; tb, tobacco; alc, alcohol.
These data indicate that common genetic variations in DNA repair and cell cycle genes contribute toward breast cancer risk. In addition, different predisposition was observed amongst BQC and NBQC breast cancer patients rendering dissimilar susceptibility toward breast cancer. The elevated risk for breast cancer in BQC may be attributable to betel quid carcinogens and minor roles of BRCA2 mutation and C allele of RAD51. Whereas NBQC could be at slightly lower risk for breast cancer due to the protection offered by the Pro/Pro-TP53 form. CCND1 polymorphism conferred protection irrespective of the betel quid chewing status.
Genomic Alterations in Breast Cancer Patients in Betel Quid and Non Betel Quid Chewers
Tobacco chewing with BQ results in increased exposure (1,000 mg/day) to carcinogenic tobacco-specific nitrosamines (TSNAs). N’-nitrosonornicotine (NNN), 4- (N-methyl-N-nitrosamino)-1- (3-pyridyl)-1-butanone (NNK). Nnitrosoanabasine (NAB), N-nitrosodimethylamine, and N-nitrosodiethylamine have been detected in saliva of BQ with tobacco chewers37 and are known to induce mammary tumors in rodents and anaphase bridges via DNA double stranded breaks causing genomic imbalances in human cells.36,38 Examination of genomic alteration due to tobacco carcinogens depicts gain on chromosomes 6 and 8, and losses on chromosomes 11 and 14 in mouse lung adenocarcinomas39 and gains of 1p and 3q in patients with tobacco exposure history in head and neck squamous cell carcinomas.40 Although the literature suggests role of BQ carcinogens in mediating genomic alterations, there is no cause and effect evidence suggesting its role in breast carcinogenesis.
To elucidate the role of betel quid carcinogens in breast carcinogenesis, we investigated genomic alterations in breast cancer patients from North East India with and without BQ chewing history. Study of copy number variations and pathway networks using SNP array and ingenuity pathway analysis (IPA) was done in 26 BQC and 17 NBQC breast cancer patients.41 BQC tumors showed significantly (p < 0.01) higher number of alterations, as compared with NBQC tumors, 48 ± 17% versus 32 ± 25 respectively. Incidence of gain in fragile sites in BQC tumors were significantly higher (p < 0.001) as compared with NBQC tumors. The chromosomal regions 7q33 and 21q22.13 were significantly (p < 0.05) associated with BQC tumors, while regions 19p13.3–19p12 and 20q11.22 were significantly associated with NBQC tumors. Gene ontology (GO) and network analysis showed that genes associated with BQC regions, 7q33 and 21q22.1 were enriched for oxidoreductase (p < 0.001) and aldo-ketoreductase activity (p = 0.015) in contrast to G-protein coupled receptor protein signaling pathway (p = 0.005) and cell surface receptor linked signal transduction (p = 0.012) for 19p13.3–19p12 and 20q11.22 regions associated with NBQC. IPA analysis for BQC associated regions revealed one top network (score 20) “drug metabolism, molecular transport, nucleic acid metabolism” encompassing genes like AKR1B1, AKR1B10, AKR1B15, ERG, ETS2 (►Fig. 6A). IPA analysis for NBQC genes revealed two top networks (score 29) “molecular transport, nucleic acid metabolism, small molecule biochemistry” and “cellular development, embryonic development, organismal development” (►Fig. 6B) encompassing genes like RPN2, EMR3, BLCAP and VAV1, NNAT and MUC16, respectively.

- (A) BQC Network 1 drug metabolism, molecular transport, nucleic acid metabolism. (B) NBQC networks: molecular transport, nucleic acid metabolism, small molecule biochemistry (Network 1) and cellular development, embryonic development, organismal development (Network 2).
Twenty-seven common regions of gain were illustrated between BQC and NBQC tumors. Both groups exhibited gain on chromosomes 1q, 5p, 7p, 8q, 12q, 16p, 17q, and 20q. Enrichment and IPA of genes associated with these regions show activation of protein kinase activity (p = 0.009) and cell junction (p = 0.01). IPA analysis revealed three top networks, (►Fig. 7). Network 1 functions in cellular movement, connective tissue development and function, cellular assembly and organization (score 43) with key role played by PTK2. Network 2 functions in cell-to-cell signaling and interaction, tissue development, organismal injury and abnormalities (score 43) with RPN2, EMR3, VAV1, NNAT, and MUC16 important genes. Network 3 functions in cell morphology, cellular assembly and organization, and cellular compromise (score 32) with key roles played by MYC and YWHAZ. Among all the tumor associated canonical pathways enriched were GNRH signaling (p = 2.92E–04), cAMP-mediated signaling (p = 3.60E–04), protein kinase A signaling (p = 3.77E–04), CXCR4 signaling (p = 4.99E–03), molecular mechanisms of cancer (p = 8.58E–03), phospholipase C signaling (p = 1.01E–02), RAR activation (p = 3.16E–02), and ILK signaling (p = 4.21E–02).

- Common networks: cellular movement, connective tissue development and function, cellular assembly and organization (Network 1), cell-to-cell signaling and interaction, tissue development, organismal injury and abnormalities (Network 2), cell morphology, cellular assembly and organization, cellular compromise (Network 3).
The study shows that breast cancer associated with betel quid chewing exemplifies genetic alterations differing from those observed in the nonbetel quid chewer. Several genetic changes are shared in both tumor groups considered as crucial in breast cancer progression.
Predictive Tumor Markers in Breast Cancer in Indian Women
Majority of breast cancer patients in India present at locally advanced stage (IIIB, IIIA, and IIB) and standard of care for these patients is NACT followed by surgery in the form of modified radical mastectomy and three more cycles of adjuvant chemotherapy.42,43 NACT facilitates local as well as distant control of the disease and provides an in vivo chemosensitivity test for a particular regime.44 It is vital to predict response to chemotherapy to tailor the regime in a particular patient for an optimum response and to avoid chemotoxicity in a nonresponder. The prediction would help in avoiding the toxicity induced by an ineffective chemotherapeutic regime in a nonresponder and would also help in the planning of an alternate regime. Studies had been undertaken to assess the predictive role of various tumor markers viz. p-glycoprotein, p53, apoptotic markers, mismatch repair genes, androgen receptor (AR) gene, and type 1 growth factor receptor family genes in search for an ideal predictor.
Role of p-glycoprotein Expression in Predicting Response to NACT in Breast Cancer
Development of resistance to chemotherapeutic agents is a major problem and one of the mechanisms considered responsible is the expression of 170-kDa membrane glycoprotein (usually referred to as p-170 or p-glycoprotein), which is encoded by multidrug resistance (MDR1) gene.45 The expression of p-glycoprotein at initial presentation has been found to be associated with refractoriness to chemotherapy and poor outcome.46 Against this background, a prospective study was conducted by studying expression of p-glycoprotein in pretreatment biopsy of tumor tissue to ascertain whether pretreatment detection of p-glycoprotein expression could be utilized as a reliable predictor of response to NACT in patients of locally advanced breast cancer (LABC). Fifty cases of locally advanced breast cancer were subjected to trucut biopsy and the tissue samples were evaluated for expression of p-glycoprotein, estrogen receptor (ER) and progesterone receptor (PR) by immunohistochemistry (IHC).47 The response to NACT was assessed clinically and by using ultrasound after three cycles of FAC (fluorouracil, adriamycin, and cyclophosphamide) regime. The clinical response was correlated with both the pre and post chemotherapy p-glycoprotein expression. A significant correlation was observed between positive p-glycoprotein expression and poor clinical response rates (p < 0.05). Significant increase in expression of p-glycoprotein was seen after three cycles of NACT, 52% before initiation of NACT increased to 73.5% after NACT. The chemotherapy induced p-glycoprotein positivity observed in the study could possibly explain the phenomenon of acquired chemoresistance and may also serve as an intermediate end point in evaluating drug response particularly if the adjuvant therapy is planned with the same regime.
The study concluded that pretreatment p-glycoprotein expression predicts and indicates a poor clinical response to NACT.
Role of p53 and Apoptotic Markers in Predicting Response to NACT in Breast Cancer
The pathway associated with tumor cell death is mainly apoptosis or programmed cell death and chemotherapeutic drugs like DNA-damaging agents act on rapidly multiplying cells including both the tumor and the normal cells by following this pathway.48,49 This could account for both the response and toxic effects. Absence or decreased apoptosis has been found to be associated with chemo resistance. The change in expression of apoptotic markers (Bcl-2 and Bax proteins) brought about by various chemotherapeutic regimens has been used to identify drug resistance in the tumor cells.50 A prospective clinical study was conducted to assess whether chemotherapy induced toxic effects could serve as reliable predictors of apoptosis or response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. Fifty cases of locally advanced breast cancer after complete routine and metastatic work up were subjected to trucut biopsy and the tissue evaluated immunohistochemically for apoptotic markers (Bcl-2/Bax ratio).51 Three cycles of NACT using FAC regime (5-fluorouracil, adriamycin, cyclophosphamide) were given at 3 weekly intervals and patients assessed for clinical response as well as toxicity after each cycle. Modified radical mastectomy was performed in all patients 3 weeks after the last cycle and the specimen were re-evaluated for any change in the Bcl-2/Bax ratio. The clinical response, immunohistochemical response and the drug-induced toxicity were correlated and compared. Immunohistochemical response was defined as decrease in the Bcl-2/Bax ratio.
Clinical response including the reduction in the tumor size and axillary lymph node status was observed in 70% of patients and was found to be statistically significant (p < 0.0001). There were no patients in the N0 group and 29.4% of the N1 patients were down staged to N0, while 70.6% remained N1. In patients with N2 disease, 7.7% were down staged to N0 status, while 46.2% were down staged to N1 status, and 46.2% did not show any response. Change in immunohistochemical expression (IHC response) was observed in 60% patients, which was statistically significant (p = 0.008). Correlation between IHC and clinical response was also found to be statistically significant (p < 0.0001).
Acute vomiting was observed in 63.3% patients. Vomiting was found in 81% clinical responders (p = 0.002) and 78% IHC responders which was statistically significant (p = 0.04). Alopecia was observed in 86% clinical responders (p = 0.000) and 94% IHC responders (p = 0.000). Leucopenia was observed in only 14% and 17% of clinical and IHC responders respectively and was found to be an insignificant predictor of response in the present study. When multiple toxicities were correlated with the clinical and IHC response, 46.7% of patients had both acute vomiting and alopecia. 67% clinical responders (p = 0.001) had both vomiting and alopecia. 72% immunohistochemical responders (p = 0.001) had both vomiting and alopecia.
A significant positive correlation was observed between the presence of vomiting (r = +0.558), alopecia (r = +0.802) and response to NACT. A significant negative correlation was observed between the absence of side effects and poor response to NACT. The chemotherapy-induced toxicity was observed to be a cost effective and reliable predictor of response to NACT.51
Correlation of Mismatched Repair Genes with Response to NACT in Breast Cancer
The DNA mismatch repair (MMR) pathway is an important postreplicative repair process. It is involved in the maintenance of genomic stability and MMR genes have therefore been named the proofreaders of replicating DNA.52 These genes repair the replicative errors of DNA and are thus imperative for genomic stability.53
The MMR genes have been found to be involved in promoting cytotoxicity, apoptosis, p53 phosphorylation, and cell cycle arrest following exposure to exogenous DNA damaging agents. Loss of MMR function prevents the correction of replicative errors leading to instability of the genome, and can be detected by polymorphisms in microsatellites nucleotide repeat sequences scattered in whole genome. This phenomenon, known as microsatellite instability (MSI), is a hallmark of MMR dysfunction and can be used as a marker of MMR dysfunction in various malignancies.54 An alternative method for detection of MMR dysfunction is to test the expression of protein products of the MMR genes by IHC, as mutations in these genes lead to reduced or absent expression of their gene products. Correlation between losses of MMR function, histopathological, and behavioral parameters of the tumor with response to chemotherapy in breast cancers may be of value in predicting response to NACT.
Thirty-one cases of locally advanced breast carcinoma were studied to assess the correlation between MMR dysfunction, clinicopathological parameters and clinical response to NACT.55 Analysis for immunohistochemical expression for four MMR protein products MLH1, MSH2, MSH6, and PMS2 was done in the pre NACT trucut biopsy specimen and after three cycles of NACT with CAF (cyclophosphamide, adriamycin, 5-fluorouracil) regimen, in the modified radical mastectomy specimen. Seventeen patients (54.83%) showed down staging of the tumor size and axillary lymph node status in response to NACT including 2 patients with complete pathological response (p = 0.000).
A significant correlation was observed between expression of MLH1 and MSH2 with histological grade (p = 0.048 and 0.038 respectively). Tumors with decreased expression of MLH1 and MSH2 showed poor differentiation. Combination of loss of expression of MMR proteins with histological grade may serve as a predictor of aggressive behavior and poor outcome. Cases with high post NACT expression of PMS2 were poor responders to chemotherapy. MSH6 was the most frequently altered MMR gene, with loss of expression in 48% patients; high expression is associated with poor response to NACT.
Role of AR in Breast Cancer
AR belongs to a family of intracellular steroid hormone receptors that function as ligand dependent transcription factor which regulates target gene expression. The full length AR protein is a 110 kDa phosphoprotein, which mediates its physiological functions by binding to its ligand testosterone or after its conversion to 5α-dihydrotestosterone (DHT) by 5α-reductases.56-58 AR has an N-terminal transactivation domain, which contains the poly-glutamine (CAG) repeat sequence, a DNA-binding domain (DBD) having two C4 type zinc fingers, a hinge region, and a C-terminal ligand-binding domain (LBD), which gets activated upon binding to androgens. Androgen binding to C-terminal of AR leads to the dissociation of chaperone proteins and dimerization of AR leading to a conformational change whereby its nuclear localization signal (NLS) is exposed. Exposed NLS then aids in the translocation of AR to the nucleus, where it binds to androgen-response elements (AREs) present in the promoters of several target genes in a tissue-specific manner. In the nucleus, AR recruits many other proteins, such as general transcription factors and RNA polymerase to activate androgen-responsive genes.59 Traditionally, ER and PR are known to be the prominent players in the progression and development of breast cancer. Recent evidence suggests that AR antagonizes ER function and plays an antiproliferative role in ER+ breast cancers whereas it plays a significant role in facilitating tumor cell growth in an androgen-dependent manner in an ER–/AR+ breast cancers.60,61
Role of AR as Independent Predictor for Response to NACT in Locally Advanced Breast Cancers
The clinical significance of AR in breast cancer has been studied by correlating its expression with clinicopathological parameters, other steroid receptors (ER and PR), and growth factors receptors (EGFR and CD105) in 100 cases by IHC.62 Risk ratio (RR) along with 95% confidence interval (CI) was estimated to assess the strength of association between the markers and clinicopathological characteristics. Categorical principal component analysis (CATPCA) was applied to obtain new sets of linearly combined expression, for their further evaluation with clinicopathological characteristics. In 31 cases presenting with locally advanced breast cancer (LABC), the expression of AR, ER, PR, EGFR, and CD105 correlated with response to NACT. The results indicate the association of AR+ (p = 0.001) and AR+/EGFR- (p = 0.001) with the therapeutic response to NACT in LABC patients. The AR expression exhibited maximum sensitivity, specificity and likelihood ratio of positive and negative test for response to NACT.
Correlation of expression of steroid (AR) and type 1 growth receptor genes (EGFR, ERBB2, and ERBB3) and gene associated with metabolism of chemotherapeutic drugs (MDR1), before and after therapy, was studied in LABC patients with response to treatment.63 Significant correlation was found with high levels of pre NACT AR gene expression (p = 0.016) with responders, which decreased following NACT (p = 0.008). Hence, AR upregulation can serve as a useful predictive marker for response to NACT. A significant posttherapeutic increase in the expression levels of EGFR and MDR1 gene in responders (p = 0.026 and p < 0.001) as well as in nonresponders (p = 0.055 and p = 0.001) suggests that expression of these genes changes during therapy but they do not have any impact on tumor response, whereas a post-therapeutic reduction was observed in AR in responders. The study indicated an independent predictive role of AR with response to NACT.
Study of AR Signaling in Breast Cancer
AR has been implicated in the development and progression of breast and prostate cancers. Therefore, understanding the regulatory mechanisms of the functioning of AR in ER-/AR+ breast cancer will provide many novel targets for the purpose of therapeutic intervention.
To get the molecular insights into AR signaling in breast cancer.64 We identified 75 AR targets having prominent roles in cell cycle, apoptosis, and metabolism by using bioinformatics tools and validated 10 genes as AR targets by studying the regulation of these genes in MDA-MB-453 cell line on stimulation by androgens like 5α-DHT using RT-qPCR and ChIP assay. Our experimental data showed that treatment of breast cancer cells with DHT promotes cell proliferation and decreases apoptosis while treatment with antiandrogen like bicalutamide was able to reverse the effect of DHT, thereby demonstrating the application of bicalutamide along with taxanes as novel therapy for the treatment of triple-negative breast cancers (TNBCs), which are positive for downstream AR signaling (►Fig. 8). To understand how AR influences DNA binding, an exhaustive, rigid-body docking between individual ARE and DNA binding domain (DBD) of the AR protein was performed. To decipher the DNA–protein contact points between the AR-DBD and ARE(s), five genes namely, AATK, CDT1, KLF6, LIPH, and SGOL2 were selected on the basis of TF search score and fold enrichment in ChIP-qPCR experiments for the purpose of interaction studies. Our data showed that novel residues K567, K588, R591, and K592 are involved in the process of DNA binding and make important contributions in increasing the stability of DNA–protein complexes. To verify these specific DNA–protein interactions, electrostatic energy term calculations for each residue were determined using the linearized Poisson–Boltzmann equation.

- Proposed model shows AR regulation of the breast tumor progression. On DHT stimulation AR translocates into the nucleus and bind to its cognate androgen response elements (AREs). Inside the nucleus, AR upregulates the expression of ABL1, CDT1, KLF6, and SGOL2, while it represses MAD1L1 expression to induce cell proliferation. At the same it induces AATK expression and down regulates the expression of BOK, BIK, and ENDOG to decrease apoptosis and promote breast cancer progression. Bicalutamide (BIC) reverses the effect of AR on cell cycle and apoptosis by binding and preventing its activation. Due to its ability to negate the effects of AR, bicalutamide can be used to block breast cancer progression.
Conclusion
These studies showed the occurrence of breast cancer at younger age in Indian women with significant annual percentage increase in incidence at younger age. The significant number of cases showing family history indicates genetic component playing a major role in the etiology. This study suggests one of the several possibilities with respect to genetic predisposition in the North Indian population. First, there may be a significant proportion of BRCA1/2 mutations that are large germ line rearrangements, which would not have been detected by the method of mutation screening employed. Second, it is possible that there are some unknown genes, which may contribute more significantly to familial breast carcinoma in this population than do BRCA1 and BRCA2.CYP17 A2 allele gene polymorphism plays a significant role for development of breast cancer in Indian women at young age and can also serve as a target for therapy. Due to association of A2 allele of CYP17 gene with increased estrogen level, the steroid hydroxylases are turning out to be potential new targets for drug development. Instead of inhibiting the action of steroid hormones at the level of their receptors by using antihormones, CYPs may help to develop new inhibitors of hydroxylase enzymes.
Betel quid chewing has been shown as a significant and independent risk factor for developing breast cancer in NE region. Although common genetic variations in DNA repair and cell cycle genes contribute toward breast cancer risk, BQC are at an elevated risk for breast cancer attributable to betel quid carcinogens with minor roles of BRCA2 mutation and C allele of RAD51. Breast carcinogenesis associated with betel quid chewing show genetic alterations different from those observed in the nonbetel quid chewers. Several genetic changes are shared in both tumor groups considered as crucial in breast cancer progression.
Expression of p-glycoprotein in pretreatment biopsy predicts a poor clinical response to NACT in patients having LABC. The chemotherapy-induced toxicity (vomiting and alopecia) correlated significantly with clinical and immunohistochemical response (reduction in Bcl2/Bax ratio) and were found to be a cost-effective and reliable predictor of response to NACT. Alteration in expression of MMR proteins was found mainly in high histological grades and predicts aggressive behavior and poor response to NACT in patients with LABC. Androgen receptor has been identified as independent predictive marker for response to NACT in locally advanced breast cancer cases. It can serve as novel therapeutic target for TNBCs. Bicalutamide (anti androgen) can be used along with other chemotherapeutic drugs as novel therapy for the treatment of TNBCs, which are positive for downstream AR signaling.
Note
Achanta Lakshmipathi Oration delivered during NAMSCON 2018 at the Mahatma Gandhi Medical College and Research Institute, Puducherry.
Conflict of Interest
None declared.
References
- Three Year Report of Population Based Cancer Registries 2012–2014. Bangalore: Indian Council of Medical Research (ICMR) 2016
- [Google Scholar]
- Changing trends in incidence of breast cancer: Indian scenario . Indian J. Cancer. 2009;46(01):73-74.
- [CrossRef] [PubMed] [Google Scholar]
- Clinico-morphological patterns of breast cancer including family history in a New Delhi hospital, India–a cross-sectional study. World J Surg Oncol. 2005;3:67.
- [CrossRef] [PubMed] [Google Scholar]
- Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684-1689.
- [CrossRef] [PubMed] [Google Scholar]
- Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088-2090.
- [CrossRef] [PubMed] [Google Scholar]
- A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66-71.
- [Google Scholar]
- Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789-792.
- [CrossRef] [PubMed] [Google Scholar]
- The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12(03):333-337.
- [CrossRef] [PubMed] [Google Scholar]
- Contribution of germline BRCA1 and BRCA2 sequence alterations to breast cancer in Northern India. BMC Med Genet. 2006;7:75.
- [CrossRef] [PubMed] [Google Scholar]
- BRCA1 and BRCA2 in Indian breast cancer patients. Hum Mutat. 2002;20(06):473-474.
- [CrossRef] [PubMed] [Google Scholar]
- Estrogen use and cancer incidence: a review. Cancer Invest. 1995;13(03):287-295.
- [CrossRef] [PubMed] [Google Scholar]
- Prevention of cancers of the breast, endometrium and ovary. Oncogene. 2004;23(38):6379-6391.
- [CrossRef] [PubMed] [Google Scholar]
- Estrogens and breast cancer. Carcinogenesis. 1996;17(11):2279-2284.
- [CrossRef] [PubMed] [Google Scholar]
- Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43(08):779-804.
- [CrossRef] [PubMed] [Google Scholar]
- Tissue-specific, cyclic adenosine 3′,5′-monophosphate-induced, and phorbol ester-repressed transcription from the human P450c17 promoter in mouse cells. Mol Endocrinol. 1990;4(12):1972-1979.
- [CrossRef] [PubMed] [Google Scholar]
- Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet. 1994;3(10):1873-1876.
- [CrossRef] [PubMed] [Google Scholar]
- Cytochrome P450c17α gene (CYP17) polymorphism is associated with serum estrogen and progesterone concentrations. Cancer Res. 1998;58(04):585-587.
- [Google Scholar]
- CYP17 gene polymorphism and its association with high-risk north Indian breast cancer patients. J Hum Genet. 2007;52(02):159-165.
- [CrossRef] [PubMed] [Google Scholar]
- Effects of synthetic vitamin D analogues on breast cancer cell proliferation in vivo and in vitro. Biochem Pharmacol. 1992;44(04):693-702.
- [CrossRef] [PubMed] [Google Scholar]
- 1 α, 25-dihydroxyvitamin D3 inhibits the invasive potential of human breast cancer cells in vitro. Clin Exp Metastasis. 1994;12(03):195-202.
- [CrossRef] [PubMed] [Google Scholar]
- Effects of 1,25 dihydroxyvitamin D3 and its analogues on induction of apoptosis in breast cancer cells. J Steroid Biochem Mol Biol. 1996;58(04):395-401.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D receptor gene polymorphism(s) and breast cancer risk in north Indians. Cancer Detect Prev. 2009;32((05) (06)):386-394.
- [CrossRef] [PubMed] [Google Scholar]
- Androgens and androgen receptors in breast cancer. J Steroid Biochem Mol Biol. 2007;105((01) (05)):1-15.
- [Google Scholar]
- Androgen receptor expresion in breast cancer: relationship with clinicopathological characteristics of the tumors, prognosis, and expression of metalloproteases and their inhibitors. BMC Cancer. 2008;8:149.
- [CrossRef] [PubMed] [Google Scholar]
- The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 1994;22(15):3181-3186.
- [CrossRef] [PubMed] [Google Scholar]
- Role of an androgen receptor gene polymorphism in development of hormone refractory prostate cancer in Indian population. Asian Pac J Cancer Prev. 2007;8(02):275-278.
- [Google Scholar]
- Is reduced CAG repeat length in androgen receptor gene associated with risk of prostate cancer in Indian population? Clin Genet. 2005;68(01):55-60.
- [CrossRef] [PubMed] [Google Scholar]
- Androgen receptor status predicts response to chemotherapy, not risk of breast cancer in Indian women. World J Surg Oncol. 2010;8:64.
- [CrossRef] [PubMed] [Google Scholar]
- Cigarette smoking and the risk of breast cancer in women: a review of the literature. Cancer Epidemiol Biomarkers Prev. 2002;11((10) Pt 1)):953-971.
- [Google Scholar]
- India has some of the highest cancer rates in the world. BMJ. 2005;330(7485):215.
- [CrossRef] [PubMed] [Google Scholar]
- Mammary expression of xenobiotic metabolizing enzymes and their potential role in breast cancer. Cancer Res. 2000;60(17):4667-4677.
- [Google Scholar]
- Smokeless tobacco and cancer. Lancet Oncol. 2008;9(07):667-675.
- [CrossRef] [PubMed] [Google Scholar]
- A case-only analysis of the interaction between N-acetyltransferase 2 haplotypes and tobacco smoke in breast cancer etiology. Breast Cancer Res. 2005;7(03):R385-R393.
- [CrossRef] [PubMed] [Google Scholar]
- Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: tobacco exposure as a risk modulator. Carcinogenesis. 2007;28(07):1455-1462.
- [CrossRef] [PubMed] [Google Scholar]
- Betel quid chewing as an environmental risk factor for breast cancer. Mutat Res. 2010;703(02):143-148.
- [CrossRef] [PubMed] [Google Scholar]
- Cigarette smoke condensate-induced transformation of normal human breast epithelial cells in vitro. Oncogene. 2004;23(35):5880-5889.
- [CrossRef] [PubMed] [Google Scholar]
- Alert for an epidemic of oral cancer due to use of the betel quid substitutes gutkha and pan masala: a review of agents and causative mechanisms. Mutagenesis. 2004;19(04):251-262.
- [CrossRef] [PubMed] [Google Scholar]
- Cigarette smoke induces anaphase bridges and genomic imbalances in normal cells. Mutat Res. 2004;554((01) (02)):375-385.
- [CrossRef] [PubMed] [Google Scholar]
- Array CGH analysis reveals chromosomal aberrations in mouse lung adenocarcinomas induced by the human lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Biochem Biophys Res Commun. 2006;341(03):856-863.
- [CrossRef] [PubMed] [Google Scholar]
- Chromosomal aberrations in patients with head and neck squamous cell carcinoma do not vary based on severity of tobacco/alcohol exposure. BMC Genet. 2002;3:22.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic alterations in breast cancer patients in betel quid and non betel quid chewers. PLoS One. 2012;7(08):e43789.
- [CrossRef] [PubMed] [Google Scholar]
- Randomized trial comparing neo-adjuvant versus adjuvant chemotherapy in operable locally advanced breast cancer (T4b N0-2 M0) J Surg Oncol. 2003;84(04):192-197.
- [CrossRef] [PubMed] [Google Scholar]
- Neoadjuvant chemotherapy in breast cancer. Br J Surg. 2005;92(01):14-23.
- [CrossRef] [PubMed] [Google Scholar]
- Primary chemotherapy in breast cancer: The beginning of the end or the end of the beginning for the surgical oncologist? World J Surg Oncol. 2003;1(01):14.
- [CrossRef] [PubMed] [Google Scholar]
- P-glycoprotein expression in treated and untreated human breast cancer. Br J Cancer. 1989;60(06):815-818.
- [CrossRef] [PubMed] [Google Scholar]
- Role of p-glycoprotein expression in predicting response to neoadjuvant chemotherapy in breast cancer–a prospective clinical study. World J Surg Oncol. 2005;3:61.
- [CrossRef] [PubMed] [Google Scholar]
- Reduced apoptosis and proliferation and increased Bcl-2 in residual breast cancer following preoperative chemotherapy. Breast Cancer Res Treat. 1998;48(02):107-116.
- [CrossRef] [PubMed] [Google Scholar]
- Apoptosis . Its significance in cancer and cancer therapy. Cancer. 1994;73(08):2013-2026.
- [CrossRef] [PubMed] [Google Scholar]
- Changes of biological features in breast cancer cells determined by primary chemotherapy. Breast Cancer Res Treat. 1997;44(03):185-192.
- [CrossRef] [PubMed] [Google Scholar]
- Is drug-induced toxicity a good predictor of response to neo-adjuvant chemotherapy in patients with breast cancer?–a prospective clinical study. BMC Cancer. 2004;4:48.
- [CrossRef] [PubMed] [Google Scholar]
- Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91(04):467-477.
- [CrossRef] [PubMed] [Google Scholar]
- Selective recognition of a cisplatin-DNA adduct by human mismatch repair proteins. Nucleic Acids Res. 1997;25(03):491-496.
- [CrossRef] [PubMed] [Google Scholar]
- The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4(01):1-6.
- [Google Scholar]
- Immunohistochemical pattern of hMSH2/hMLH1 in familial and sporadic colorectal, gastric, endometrial and ovarian carcinomas with instability in microsatellite sequences. Virchows Arch. 2001;438(01):39-48.
- [CrossRef] [PubMed] [Google Scholar]
- Repressive effects of resveratrol on androgen receptor transcriptional activity. PLoS One. 2009;4(10):e7398.
- [CrossRef] [PubMed] [Google Scholar]
- Prostate cancer schemes for androgen escape. Nat Med. 2000;6(06):628-629.
- [CrossRef] [PubMed] [Google Scholar]
- Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(03):601-610.
- [CrossRef] [PubMed] [Google Scholar]
- Regulation of the androgen receptor by post-translational modifications. J Endocrinol. 2012;215(02):221-237.
- [CrossRef] [PubMed] [Google Scholar]
- An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25(28):3994-4008.
- [CrossRef] [PubMed] [Google Scholar]
- Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol. 2012;26(08):1252-1267.
- [CrossRef] [PubMed] [Google Scholar]
- Expression of androgen receptor in breast cancer & its correlation with other steroid receptors & growth factors. Indian J Med Res. 2012;135(06):843-852.
- [Google Scholar]
- Study on predictive role of AR and EGFR family genes with response to neoadjuvant chemotherapy in locally advanced breast cancer in Indian women. Med Oncol. 2012;29(02):539-546.
- [CrossRef] [PubMed] [Google Scholar]
- A molecular analysis provides novel insights into androgen receptor signalling in breast cancer. PLoS One. 2015;10(03):e0120622.
- [CrossRef] [PubMed] [Google Scholar]





