Translate this page into:
Detection of Antibiotic Resistance and Biofilm- Producing Ability of Staphylococcus Species in Clinical Isolates
Address for correspondence Minakshi Bhattacharjee, PhD, Department of Biotechnology, Assam Downtown University, Gandhinagar, Panikhaiti, Guwahati, Assam 781026, India (e-mail: minakshibhattacharjee2007@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Staphylococci are responsible for life-threatening infections in hospitals and community. Their ability to produce multiple virulence factors and antibiotic resistance is an important reason of high mortality in staphylococcal infections. Biofilm production by these organisms makes it difficult to treat. Most of the treating antibiotics are failing and making it a matter of concern.
Aims
This study aims to detect the increased antibiotic resistance in biofilm-producing Staphylococcus and to compare the performance of three potential methods of detection.
Methods
A total of 81 isolates of staphylococci including coagulase negative staphylococci (CoNs), methicillin resistant S. aureus (MRSA), and methicillin sensitive S. aureus (MSSA) are included in this study. After the identification, an antibiotic sensitivity test was performed. Biofilm detection was done by three different methods: Congo red agar method, tube adherence method, and microtiter plate method.
Result
Out of the 81 samples, 37 CoNs, 17 MRSA, and 27 MSSA were identified. Out of them we got 43 (53%) biofilm producers by Congo red agar method, 40 (49%) by tube adherence method, and 52 (64%) producers by tissue culture plate/microtiter plate method. Most of the biofilm producers showed multiple drug resistance.
Conclusion
We found out that the microtiter plate method is sensitive and reliable as compared with the other two methods. Antibiotic resistance was found to be very common in biofilm producers. This was due to the resistance developed as a result of the matrix that does not let the antibiotic bind with the organisms. This can make the treatment of Staphylococcus very difficult in the future as the rate of drug resistance is faster as compared with newly emerging antibiotics.
Keywords
biofilm
antibiotic resistance
MRSA
MSSA
CoNs
Introduction
Staphylococcus aureus is a pathogen that can infect human as well as animals. The infections can range from skin and soft tissue infection to serious life-threatening illnesses like endocarditis, septicemia, and toxin-mediated shock syndrome in human, and skin infections and mastitis caused by S. aureus are reported in animals.1 Staphylococcus aureus has many virulence factors that help the organism survive and initiate infection inside the host. These factors act in different ways to colonize, invade, and spread themselves. This is the primary reason for S. aureus being able to attack immunologically competent hosts.2 The ability of the organism to develop biofilm enhances the severity of infections caused by S. aureus as well as increases the antibiotic resistance.3 The steps of biofilm formation involve adhesion of the organism to the surface, which is aided by the expression of various microbial surface components that recognize adhesive matrix molecules. These molecules can bind to various extracellular matrix components, including elastin, fibronectin A and B, laminin, collagen, fibrinogen, and clumping factors. These proteins can connect to the cell wall or other surfaces by using similar signal sequences. They can cover medical equipment and start the creation of proteins like biofilm matrix proteins in a bacterial matrix.4,5 Several Staphylococcus species now exhibit significant virulence traits due to the production of biofilm.5 The biofilm serves as a protective cover that provides a favorable environment to the bacteria and allows them to survive against adverse conditions.6 In addition to its ability to form biofilm on medical devices, which leads to the removal of devices, S. aureus can form biofilm inside the human body. This leads to infection associated with resistance to antimicrobial agents and failure of the treatment resulting in removal of the infected tissue to control the infection.7 The resistance caused by biofilm is due to the presence of bacteria inside the biofilm; they remain dormant during the antimicrobial therapy and later emerge as resistant strains. Another mechanism that leads to resistance to antibiotics is the inability of antimicrobials to penetrate through the layers of biofilm.8 Numerous coagulase negative staphylococci (CoNs) species, including S. epidermidis, have emerged as the primary carriers of nosocomial infections, particularly those that affect newborns, people with immunological disorders, and patients receiving care in the intensive care unit who utilize catheters or other prosthetic devices.9 Catheter-associated infections may be caused by CoNs that produce biofilm that affects immunocompromised patients, including premature infants, neutropenic cancer patients, elderly people with serious underlying diseases, hospitalized patients undergoing invasive procedures, and people with permanent plastic devices.10 Methicillin resistant S. aureus (MRSA) is largely found to be resistant to common available antibiotics. One of the common reasons for high mortality and morbidity in MRSA infections are biofilm producers.11 In hospitals, the biofilm-producing pathogens are becoming a serious concern, and the existing antimicrobial agents are failing to treat these pathogens. The increase in multidrug-resistant organisms with biofilm is weakening the health care policy.12 Therefore, the development of clinically applicable biofilm experimental modalities is urgently required since biofilm plays a significant role in the slow healing of wounds.13
In our study, 81 isolates belonging to the genus Staphylococcus were investigated for the presence of biofilm forming activity by three different methods such as the tube adherence method, Congo red agar method, and tissue culture plate method. Antibiotic resistance was compared between the biofilm producers and nonproducers. We found that the multidrug resistance was common among the biofilm producers and not all the strains showed presence of biofilm by all the methods used in this study.
Materials and Methods
Sample Collection
A total of 81 strains of staphylococci isolated from various clinical samples were collected. We included organisms that were clinically significant pathogens and not contaminants or those with insignificant growth. The samples included were pus, throat swab, urine, nasal swab, tracheal secretion, ear swab, vaginal swab, etc.
Identification of Staphylococcus Species
Gram staining and other biochemical tests such as slide and tube coagulase tests, catalase test, and other identification tests were used to distinguish between different species of staphylococci. After an overnight incubation, S. aureus is recognized by its golden yellow β-hemolytic colonies on blood agar, catalase production, and coagulase production. Staphylococci that are coagulase negative are CoNs and were further identified up to the species level.14 All the S. aureus strains were further tested for the identification of MRSA using Cefoxitin disc (30 mcg) by the Kirby–Bauer disc diffusion method. The results were given as per the CLSI (Clinical laboratory Standard Institute) guidelines.15
Antibiotic Sensitivity Testing
The antibiotic sensitivity test was performed by the Kirby–Bauer disc diffusion method and interpreted based on the CLSI guidelines. The antibiotic discs tested were gentamycin (G), erythromycin (E), tetracycline (TE), ciprofloxacin (CIP), trimethoprim/sulfamethoxazole (TS), linezolid (L), teicoplanin (TE), tetracycline (TE), ampicillin (AM), cefuroxime (CXM), clindamycin (CD), levofloxacin (LV), amoxicillin-clavulanic acid, ceftazidime [CFT], nitrofurantoin [NF], tigecycline [TG] and doxycycline [DX], cefoxitin (Cx). The test was performed on Mueller–Hinton agar (Himedia India).
ATCC 25923 and ATCC 43300 were used as the controls for methicillin sensitive S. aureus (MSSA) and MRSA, respectively.15,16,17
Detection of Biofilm
The detection of biofilm production was done by three different methods to detect their efficacy and also to compare the findings along with the antibiotic resistance.
Congo Red Agar Method
The detection of biofilm by the Congo red agar method was done by using the method given in a previous study. The test organisms are inoculated into a special media prepared using brain–heart infusion medium supplemented with sucrose (5%) and Congo red dye. After the inoculation, the plates were incubated at 37°C for 24 to 48 hours. The test was interpreted positive for biofilm if the colonies were black in color with dry crystalline consistency. Weakly positive showed pink colonies with occasional black color in the center of the colonies (►Fig. 1A).18

- (A) Biofilm producers show black metallic colonies on Congo red agar. (B) Biofilm producers show material adhering to the bottom. (C) Tissue culture plate method.
Tube Adherence Method
The bacterial suspension of the test organisms was incubated at 35°C in glass test tubes containing brain–heart infusion broth for 48 hours. After the incubation was over, the supernatant was discarded and the glass tube was stained with 0.1% of safranine solution. The tube was washed three times with distilled water and then air dried. Any material adhering to the test tube surface was considered as positive and presence of only a ring was considered negative (►Fig. 1B).19
Microtiter Plate Method/Tissue Culture Plate Method
The detection of biofilm was also done by the tissue culture plate method using the procedure given by Christensen et al in 1995.20 Fresh growths of the test isolates were inoculated into 5 mL of Trypticase soy broth and then incubated for 24 hours at 37°C. After 24 hours of incubation, the culture was further diluted to 1:100 using fresh Trypticase soy broth. A 96-well polystyrene plate was used; each well was filled with 0.2 mL of the bacterial inoculums and incubated for 24 hours at 37°C. After the incubation the bacterial suspension was removed from all the wells by gently tapping the plate. The wells were washed two times using phosphate-buffered saline (pH 7.2) and then allowed to incubate for 1 hour at 37°C. After that the wells were stained for 10 minutes using 0.2 mL of Crystal violet (0.1%).21 Any excess amount of stain is removed by washing with distilled water twice. After washing the plates were kept to dry. Then 200 µL of glacial acetic acid was added to each well and then the optical density of the isolate was determined by an ELISA Microtiter plate reader (Alere AM2100) at 570 nm wavelength and the interpretation of the result was done as strong, moderate, and weak or nonbiofilm producers (►Fig. 1C). This interpretation was based on a previous study conducted by Stepanović et al (►Table 1).22
| OD value (average) | Biofilm production |
|---|---|
| <0.17 | Negative |
| 0.17–0.34 | Weak positive |
| 0.35–0.68 | Moderate positive |
| >0.68 | Strong positive |
Result
In this study we included 81 isolates of Staphylococcus isolated from various clinical samples. The distribution of S. aureus and CoNs among the different types of samples is shown in ►Fig. 1. Of the 81 samples, S. aureus was isolated from 44 (54%) samples and CoNs from 37 (45.6%) samples. Staphylococcus aureus was mostly isolated from pus (n = 17) and CoNs were present mostly in the nasal swabs (n = 17).
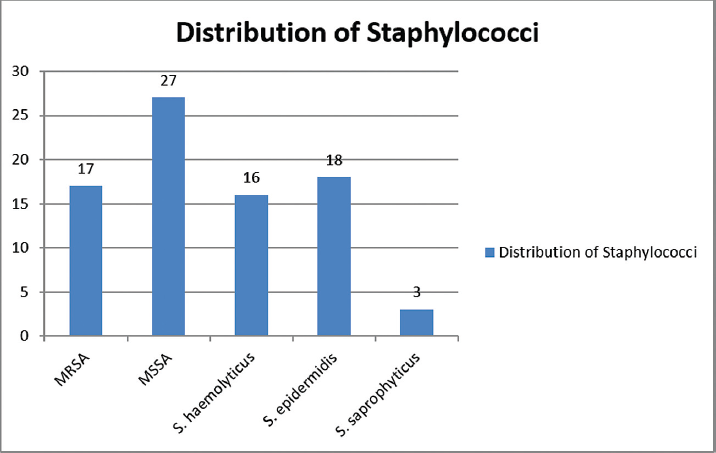
Out of the 44 S. aureus found 27 were MSSA and 17 were MRSA and out of the total 37 CoNs, 18 were identified as S. epidermidis, 16 as S. haemolyticus and 3 as S. saprophyticus (►Fig. 2).
- Distribution of staphylococci isolated from various clinical samples.
Antibiotic Resistance Pattern
The antibiotic resistance pattern of the isolates was determined by Kirby–Bauer disc diffusion method. The resistance pattern of the isolates against antibiotics is given in ►Table 2. Many strains of MRSA, MSSA, and CoNs showed multiple drug resistance.
| Cefoxitin | Benzylpenicillin | Ampicillin | Amoxicillin/clavulanic Acid | Cefuroxime | Gentamicin | Ciprofloxacin | Levofloxacin | Erythromycin | Clindamycin | Linezolid | Daptomycin | Teicoplanin | Doxycycline | Tetracycline | Tigecycline | Nitrofurantoin | Trimethoprim/sulfamethoxazole | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRSA (17) | 17 | 16 | 17 | 17 | 17 | 6 | 10 | 10 | 12 | 5 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 7 |
| MSSA (27) | 0 | 23 | 25 | 2 | 3 | 3 | 22 | 23 | 18 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| CoNs (37) | 19 | 24 | 4 | 3 | 3 | 3 | 17 | 17 | 14 | 23 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 10 |
Abbreviations: CoNs, coagulase negative staphylococci; MRSA, methicillin-resistant S. aureus; MSSA, methicillin sensitive S. aureus.
Biofilm Formation
The biofilm detection of all the strains was done by total three methods. The findings of presence of biofilm by different methods are given in ►Table 3.
| No. of isolates (N = 81) | CoNs (37) | MRSA (17) | MSSA (27) | Total positive | Total negative |
|---|---|---|---|---|---|
| Congo red agar | 18 | 9 | 16 | 43 | 38 |
| Tube adherence | 15 | 10 | 15 | 40 | 41 |
| Tissue culture plate method | 22 | 13 | 17 | 52 | 29 |
| Positive by all the methods | 13 | 9 | 13 | 35 |
Abbreviations: CoNs, coagulase negative staphylococci; MRSA, methicillin-resistant S. aureus; MSSA, methicillin sensitive S. aureus.
The table above shows the numbers of strains showing biofilm production by different methods used. All the 81 strains of S. aureus were subjected to testing for biofilm by three methods. The tissue culture plate method was one of the most promising of the three methods, since it showed a large number of isolates (n = 52) that are positive for biofilm followed by Congo red agar (n = 43), and tube adherence (40). Many strains showed positive by one or two methods only. Only 13 CoNs, 9 MRSA, and 13 MSSA were positive by all the three methods used in this study.
Discussion
Staphylococcus aureus is present on the skin and mucous membrane of the nasal cavity of human beings. It can cause multiple infections like septicemia, pneumonia, urinary tract infection, and skin infections in human. Production of biofilm by them is an important virulence factor that allows the bacteria to adapt to the adverse conditions and difficult to treat. This is also responsible for antimicrobial resistance. This calls for the need of removal of biofilm-producing organisms from medical devices and for the detection of biofilm-producing ability of the organism.21
In our study, we included 81 isolates of Staphylococcus species. The isolates were tested for their ability to produce biofilm as a virulence factor by three different methods. With regard to these three techniques, the Congo red agar method detected 43 (53%) biofilm producers among the isolates and 38 nonproducers; the tube adherence method detected 40 (49%) positive biofilm detections and 41 nonproducers, and the tissue culture plate method detected 52 (64%) producers and 29 nonproducers. Looking at the positivity rate given by various methods, the microtiter plate method is highly efficient. In a study conducted by Amita Jain in 2009, the microtiter plate method is used as the gold standard test for biofilm to evaluate the sensitivity and specificity of Congo red agar method.23 Tube adherence on the other hand has given the least rate of positivity; this could be due to weak biofilm producers which fail to adhere well to the tube. Most of the biofilm positive strains showed resistance to multiple antibiotics. CoNs have shown resistance against many antibiotics like clindamycin, levofloxacin, and trimethoprim-sulfamethoxazole. MSSA and MRSA also showed significant degrees of resistance to antibiotics like ciprofloxacin, erythromycin, and amoxicillin- clavulanic acid.
Conclusion
Clinically isolated strains of S. aureus recovered from infection in hospitalized patients show high levels of biofilm formation. Higher rates of antibiotic resistance were demonstrated by biofilm manufacturers compared with nonbiofilm manufacturers. The biofilm formation of S. aureus, MRSA, and CoNs can lead to high rates of antibiotic resistance, making treatment difficult. Biofilm screening must be done to prevent it. This study detects the phenotypic ability of Staphylococcus species to produce biofilm; a genotypic detection of associated genes can give better results.
Conflict of Interest
None declared.
Funding
None.
References
- Inhibition of Staphylococcus aureus biofilm formation by gurmarin, a plant-derived cyclic peptide. Front Cell Infect Microbiol. 2022;12:1017545.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial treatment of Staphylococcus aureus biofilms. Antibiotics (Basel). 2023;12(01):87.
- [CrossRef] [PubMed] [Google Scholar]
- Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(06):1838-1850.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus and biofilms. Mol Microbiol. 2002;43(06):1367-1378.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg Infect Control. 2016;11:Doc07.
- [Google Scholar]
- A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics (Basel). 2022;12(01):12.
- [CrossRef] [PubMed] [Google Scholar]
- Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4(12):e01067.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int J Environ Res Public Health. 2021;18(14):7602.
- [CrossRef] [PubMed] [Google Scholar]
- The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One. 2012;7(07):e40041.
- [CrossRef] [PubMed] [Google Scholar]
- Bacterial biofilms with emphasis on coagulase-negative staphylococci. J Venom Anim Toxins incl Trop Dis. 2008;14(04):572-596.
- [CrossRef] [Google Scholar]
- In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25-32.
- [CrossRef] [PubMed] [Google Scholar]
- Biofilm formation by pathogens causing ventilator-associated pneumonia at intensive care units in a tertiary care hospital: an armor for refuge. BioMed Res Int. 2021;2021:8817700.
- [CrossRef] [PubMed] [Google Scholar]
- Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci Rep. 2018;8(01):9431.
- [CrossRef] [PubMed] [Google Scholar]
- Gram Positive cocci: part-1: Staphylococci & related Gram positive cocci. Koneman's Color Atlas and Textbook of Diagnostic Microbiology 2006:645-648.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100 2020
- [Google Scholar]
- Antibiotic susceptibility pattern and biofilm formation in coagulase negative staphylococci. J Infect Dev Ctries 2014–. 1645;8(12):1643-1645.
- [CrossRef] [PubMed] [Google Scholar]
- In vitro biofilm formation and antibiotic susceptibility patterns of bacteria from suspected external eye infected patients attending ophthalmology clinic, Southwest Ethiopia. Int J Microbiol. 2020;2020:8472395.
- [CrossRef] [PubMed] [Google Scholar]
- New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42(08):872-874.
- [CrossRef] [PubMed] [Google Scholar]
- Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37(01):318-326.
- [CrossRef] [PubMed] [Google Scholar]
- Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(06):996-1006.
- [CrossRef] [PubMed] [Google Scholar]
- Difference in biofilm development capability of vancomycin and ciprofloxacin resistant Staphylococcus aureus clinical isolates. Res J Infect Dis 2013:1-8.
- [CrossRef] [Google Scholar]
- Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Acta Pathol Microbiol Scand Suppl. 2007;115(08):891-899.
- [CrossRef] [PubMed] [Google Scholar]
- Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Methods. 2009;76(01):88-92.
- [CrossRef] [PubMed] [Google Scholar]





