Translate this page into:
SARS-CoV-2 Genome Structure, Pathogenesis, Issues, and Challenges in Laboratory Diagnosis
Address for correspondence Gopal Nath, MBBS, MD, PhD, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, 221005, India (e-mail: gopalnath@gmail.com).
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Severe acute respiratory syndrome coronavirus 2 causing coronavirus disease 2019 pandemic disease is an enveloped virus, showing genome similarity with bat coronavirus. This virus initially infects the upper respiratory tract, with subsequent spread to the lower respiratory tract. Despite the availability of antigen and antibody detection methods, reverse transcription-polymerase chain reaction (RT-PCR) is the diagnostic test of choice for this novel coronaviral infection. Care must be taken while interpreting the RT-PCR results, as single RT-PCR, especially in early days of infection, maybe false negative. The availability of cartridge-based nucleic acid amplification test has improved the diagnostic facilities in a peripheral setting of developing countries.
Keywords
SARS-CoV-2
COVID-19
RT-PCR
TrueNAT
antigen
Introduction
The members of the Coronaviridae family are associated with human and animal infections, mostly affecting the respiratory and intestinal systems.1 In the past, severe acute respiratory syndrome (SARS) was reported to be caused by coronavirus (CoV-1), now designated as SARS-CoV-1. This virus claimed many lives in China in 2002. Almost a decade later, in 2013, another coronavirus named the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) emerged and caused infection in Middle Eastern countries. The present novel coronavirus (SARS-CoV-2) outbreak, though reported to the World Health Organization (WHO) on December 31, 2019, by Chinese health officials, could be traced back to November 17, 2019. It has been linked to a seafood market in Wuhan, Hubei Province, China. As per the WHO convention held on February 17, 2020, infection/disease caused by SARS-CoV-2 has been named coronavirus disease-19 or COVID-19.2 Outside of China, on January 13, 2020, the first case of SARS-CoV-2 was reported from Thailand, whereas in the United States, the first case was detected on January 20, 2020. In India, the first case of COVID-19 was reported on January 30, 2020, in Thrissur city of Kerala in a student who returned from Wuhan city of China. Subsequently, India declared a complete lockdown on March 23, 2020, to contain the spread of COVID-19. In the fourth week of January 2022, the total global case reported reached 370 million, with5.65 million deaths.3 In India, the Indian Council of Medical Research (ICMR) data shows a total of 40,858,241cases with 4,93,218 deaths.4 Early diagnosis and physical isolation are the mainstays for containing any outbreak/epidemic. This review primarily focuses on the virological aspect of SARS-CoV-2/COVID-19, its pathogenesis, and immunological response concerning knowledge and progress in the diagnostic strategies.
Genome Structure of SARS-CoV-2
The family of Coronaviridae is comprised of positive-sense, single-stranded RNA (genome of size 26–32 kb) viruses with a diameter of 80 to 120 nm. There are four coronavirus genera: α-CoVs, β-CoVs, γ-CoVsv, and δ-CoVs. Human CoVs belong to α-CoV (HCoV-229E and NL63) and β-CoVs (MERS-CoV, SARS-CoV, HCoV43, and HCoV-HKU1).4 The whole-genome sequencing of the novel CoV-2 (nCoV-2) revealed that it differs from the previously known human β-CoVs.5 The nCoV-2 strain detected elsewhere showed 99.6% sequence similarities. However, it has only 50% sequence similarity with MERS-CoV. On the other hand, this new coronavirus shows 88% identity to the bat-derived SARS-CoVs: bat-SL-CoVZC45, and bat-SLCoVXC21. Therefore, the novel β-CoV is named SARS-CoV-2 by the International Virus Classification Commission.
SARS-CoV-2 is a spherical enveloped virus particle. The envelope bears club-shaped glycoprotein spikes. The nucleic acid RNA is associated with a nucleoprotein with a capsid comprised of matrix protein (►Fig. 1). Few members of Coronaviridae contain a hemagglutinin-esterase protein (HE).6 Genome size of SARS-CoV-2, sequenced recently, is approximately 29.9 kb.7 A typical CoV consists of at least six open reading frames (ORFs) in its genome. The genes for major structural proteins in coronavirus occur in the 5-3 order. These genes are spike (S), envelope (E), membrane (M), and nucleocapsid (N).8 The first ORFs (ORF1a/b) cover almost two-thirds of the whole genome length and encode 16 NSPs (nonstructural proteins). Frame shift mutation between ORF1a and ORF1b produces two polypeptides: pp1a and pp1b. These polypeptides are further processed by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro) and one or two papain-like proteases into 16 NSPs. The single-guide RNA (SgRNA) of CoVs leads to the translation of all structural and accessory proteins (►Fig. 2). All the four major structural proteins, S, M, E and N, are encoded by the ORFs near the 3 terminus.8 Many accessory proteins, both structural and non-structural such as HE protein, 3a/protein and 4b/b protein, carry out essential functions in genome maintenance and virus replication.9
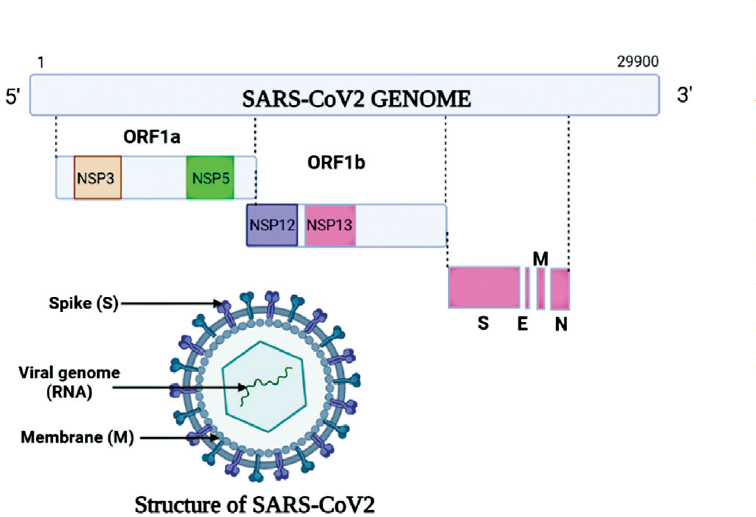
- Structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. E, envelope; M, membrane; N, nucleocapsid; NSP12, nonstructural protein 12; ORF1a, open reading frame 1a; S, spike.
- Genomic structure of severe acute respiratory syndrome coronavirus 2 virus.
The most abundant viral protein is M glycoprotein. Of the two ends of this M glycoprotein, short NH2 domain peeps outside the virus and a COOH terminus remain inside the virion.6 The other inducer of neutralizing antibodies is the S protein, a type I membrane glycoprotein that constitutes the peplomers. Spike protein is probably responsible for molecular interaction with the host receptors and viral replication. Membrane protein plays a predominant role in the intracellular assembly of virus particles independent of S protein.
The arrangement of N, E and M proteins among β-CoVs differs from other coronaviruses. The untranslated 5 and 3 regionś regions (UTRs) are involved in inter and intramolecular interactions. These UTRs are responsible for RNA–RNA interactions and binding of the viral and cellular proteins.10 The genomic size variation occurs when the first ORF at the 5 end is analyzed. Pb1ab is the first ORF of the whole genome length encoding nonstructural proteins with the size 7096aa, 7073aa, and 7078aa in SARS-CoV-2, SARS-CoV-1, and MERS-CoV, respectively. Comparative genomic analysis revealed a specific mutation pattern between human coronaviruses: SARS-CoV-2 and bat-SARSr-CoV RaTG13.11
Each of the genomes of all the SARS-CoV-2 strains submitted recently contained nearly 29,900 nucleotides (nt), which are assumed to have at least 14 ORFs (5′ to 3′), such as ORF1ab (P; 21,291 nt), spike (S; 3,822 nt), ORF3a (8,28 nt), envelope (E; 228 nt), membrane (M; 669nt), ORF8 (366 nt), and nucleocapsid (N;1,260 nt).5 The spike gene is highly divergent (93.1% nucleotide identity) when compared with that of bat-SARSr-CoV RaTG13.113.11 The spike gene encodes a glycoprotein that is crucial for determining host tropism and transmission capacity.
Pathogenesis
SARS-CoV-2 virus enters the respiratory tract through the aerosol and respiratory droplets, primarily affecting the multiciliated cells in the nasopharynx, trachea, or sustentacular cells in the nasal olfactory mucosa. The viral infection usually causes asymptomatic-to-mild symptoms in most patients. However, only a small fraction of people suffers serious consequences like pneumonia and gastrointestinal (GI) disturbances. Thus, the vicious cycle of SARS-CoV-2 pathogenesis occurs in viral entry and its multiplication, immune response, cellular damage, and recovery.12,13
SARS-CoV-2 Viral Entry and Its Multiplication
The virus enters the human body through inhalation, primarily infecting the upper respiratory tract via the acetyl choline esterase 2 (ACE2) receptor on the cell membrane. Moreover, the virus has an S(Spike) protein on its surface that mediates the target cell surface attachment, its engagement to complimentary receptors, and the membrane fusion. ACE2 receptors are present in the respiratory epithelial cells, GI, renal, cardiac tissues, and blood vessels smooth muscle cells. In addition, the binding affinity of the S protein to ACE2 receptors is much higher for SARS-CoV-2.12,13
During the initial phases, the S protein mediates the binding of the SARS-CoV-2 virus with its cellular entry on the surface. The receptor binding domain of the S1 subunit of the S protein binds with the peptidase domain of the human ACE2proteins, whereas the S2 subunit mediates membrane fusion. Subsequent binding of the S1 subunit to ACE2 and transmembrane serine protease TMPRSS2 enzyme cleaves the spike protein at the S2 site so that the S2 subunit fuses the viral and human lipid layers. This fusion results in the release of the viral nucleoprotein in the cellular cytoplasm. Subsequently, the positive sense single-stranded RNA-mediated protein synthesis occurs in the cytoplasm. In addition, the newly synthesized viral particles, assembled in the Golgi apparatus, acquire the lipid component from host cells, and form the envelope. Subsequently, the whole virion is released from the infected cells.12,13
Immune Response
Innate immune response against the viral pathogen is a major defense mechanism. During the viral replication, the pattern recognition receptor present in the cell initiates the innate immune response. STAT1, myeloid differentiation primary response protein MyD88, Toll/interleukin-1 receptor (TIR) domain-containing adapter molecule 2 in the toll-like receptor signaling pathway initiate the innate response, that result in release of interleukin-1and other proinflammatory cytokines. In addition, retinoic acid-inducible gene-I-like receptor induces the secretion of interferon. Apart from these, macrophages are crucial defense cells in viral infection. However, SARS-CoV-2 multiplies in the macrophages thus proinflammatory cytokines are released in the circulation.
In addition to the innate immune response, the adaptive immune response against SARS-CoV-2 occurs. The antigen-presenting cell targets the viral antigens to the T and B cells, resulting in the cellular and humoral immune response. Presence of raised proinflammatory T helper 17 (Th17) cells and cytotoxic CD8+ T cells in SARS-CoV-2 infected patients signifies the importance of cellular immune response. In addition, CD4 + T cells mediate the specific immunoglobulin A (IgA), IgM, and IgG humoral immune response. Briefly, the S protein of SARS-CoV-2 binds with antigen-presenting cells and binds with CD4 + T cells through Major Histocompatibility complex II (MHCII); thus, neutralizing antibodies are produced. However, these neutralizing antibodies are short-lived. The memory T and B cells sensitized with SARS-CoV-2 antigen remain for a life-time.
Activation of both innate and adaptive immune response ultimately results in the release of pro-inflammatory cytokines, such as interleukin-1 (IL-1), IL-6, IL-8 and tumor necrosis factor-alpha (TNF-α), and elevated concentrations of inflammatory markers, including D-dimer, ferritin, and C-reactive protein (CRP) at the site of infection.
Cell Damage
In a small fraction of SARS-CoV-2 infected patients, lung, kidney, heart and muscles are affected severely, where pneumonia, acute respiratory distress syndrome (ARDS), vasculitis and hemorrhage, GI manifestation, cardiac arrhythmia, and clinical manifestations of acute kidney injury occur. These manifestations are common in patients having predisposing risk factors as old age, patients having chronic obstructive pulmonary disease, cardiac, and renal issues, pregnancy, and immunocompromised patients.14
Tissue injury in SARS-CoV-2 infections occurs due to direct damage by the virus or improper immune response. The generated response against the SARS-CoV-2 virus is protective on the one side, whereas, on the opposite side, elevated immune response triggers cell damage. In addition to clearing the infected virus, the inflammatory cytokines and chemokines directly damage the cells.
During SARS-CoV-2 infection, an intense innate and adaptive immune response occurs. During this, a massive release of cytokines (storm), especially IL-6, occurs in the circulation.15 The IL-6 induces the release of acute phase reactant as CRP, hepcidin, serum amyloid A, and fibrinogen.16 In addition, IL-6 induces the adaptive immune response, where it stimulates the bone marrow to convert the plasma cells into the effective B cells, which subsequently produces antibodies against the specific antigen. These synthesized antibodies bind with antigen and initiate the classical pathway of complement system. During this pathway, chemokines and cytokines as c3a, c5a are released that recruit the inflammatory cells as neutrophil, macrophages at the site of action. In addition, the released c3b initiates the phagocytosis. Apart from IL-6, level of IL-1, IL-8, and TNF-α cytokines is increased, which indirectly results in tissue damage. Macrophages secret the IL-8 that recruits the neutrophil at the site of infection, that again releases the inflammatory cytokines; thus, a cascade pathway results in a massive release of cytokines, resulting in tissue damage.17
Recovery
Spontaneous recovery in SARS-CoV-2 infections usually occurs in majority of the patients. This recovery occurs due to the combined effect of clearing of the virus and inflammatory cytokines from the circulation. In addition, at the time of recovery, neutralizing antibodies are present in the circulation. However, the time for complete recovery varies, depending on the pre-existing health and diet. A small fraction of the infected patients has long-term consequences such as fibrosis, persistent inflammation, and catabolic syndrome.
Reinfection and relapse are common consequences of SARS-CoV-2 infection. In relapse, the infection occurs due to the sample strain and remains persistent in the immunologically privileged site. However, reinfection usually occurs with a mutant strain of SARS-CoV-2 such as omicron and BF7.
Mode of Transmission
SARS-CoV-2 is transmitted through direct, indirect, or close contact with infected people through infected secretions such as saliva and respiratory secretions or droplets. The virus is transmitted through particles of more than 5 μm as droplets and less than 5 μm as aerosols. These droplets remain on inanimate surfaces for long; thus, transmission can occur indirectly through touching surfaces with ultimate transmission to the mouth, nose, or eyes by contaminated hands.18
The Variant of Concern
During the viral replication, a frequent mutation occurs in the genetic code; thus, the newer generated virus has some different phenotypic and genotypic morphology from the previous one. These mutations alter the virus's properties, especially its transmission rate, the severity of the diseases, and diagnostic tools. The newer strains of the viruses are known as variant of concern (VOC) or variant of interest (VOI). VOI is the altered virus having a different phenotype and genotype, where reduced neutralization of the mutated virus is observed with the antibodies targeted against the previous infective virus as there is an alteration in the binding receptors. In contrast, VOC strains have evidence of a higher rate of viral transmission, increased disease severity, reduced effectiveness of the treatment, and diagnostic failure.19 Variant high consequence are the mutated viruses with clear evidence of reduced effectiveness of preventive/therapeutic strategies and diagnostic failure. Presently, most of the mutated strains are SARS-CoV-2 that are considered VOC.19
In India, Indian SARS-CoV-2 Genome Sequencing Consortia reported the presence of global VOCs, B.1.1.7, B.1.351, and P.1 (the alpha, beta and gamma VOCs, respectively). Between December 2020 and February 2021, the delta and kappa variants (B.1.617.2 and B.1.617.1, respectively) comprised 60% of sequences from Maharashtra. During the third wave of SARS-CoV-2 infection in India, the Omicron variant was reported to be a causative strain, which was classified as VOC by the US government SARS-CoV-2 Interagency Group on November 30, 2021.20
Laboratory Diagnosis of SARS-CoV-2: Issues and Challenges
Let us consider specific key points before we discuss in detail the laboratory diagnosis of SARS-CoV-2 infection:
It is established that COVID-19 cases are occurring because of community transmission through air droplets. The laboratory diagnosis detects the virus from the infected person in the early days of symptoms. Early diagnosis is the hallmark in containing the spread of infection further through isolation and contact tracing.21,22,23 The direct examination under the electron microscope is nonspecific and cumbersome. Isolation of the virus is risky and needs a biosafety level-3 (BSL-3) facility. In addition, antigen detection is not sensitive as the copy number cannot be increased; thus, we can only detect antigens during the symptomatic phase of COVID-19.
Moreover, antibody detection, although immune response usually amplifies the humoral and cellular responses, is not feasible as it appears late in infection. Thus, amplification of nucleic acid remains the most suitable and reliable detection method. Currently, the diagnosis is made using real-time reverse transcription-polymerase chain reaction (RT-PCR), similar to those developed for SARS/MERS-CoV.24,25 This review primarily focuses on the intricacies of diagnosis must be understood by clinicians, clinical microbiologists, and public health authorities.
In the following section, we describe the steps needed for the laboratory diagnosis of SARS-CoV-2: preanalytical, analytical, and interpretation of results.
Preanalytical Issues
Upper Respiratory Tract Infection and Collection of Samples for Diagnosis of COVID-19 Caused by SARS-CoV-2
The collection of nasopharyngeal secretions appropriately is an essential step. For virus detection, if the sample is not collected properly, it may give false-negative results. It has been reported that after the 5 to 6 days of infection, a significant viral load occurs in the upper and lower respiratory tract.26,27,28,29 The preferred method is the collection of nasopharyngeal secretions with the same swab from both nasopharynges (NP). However, another site to collect the secretions by the second swab is the oropharynx (OP). Subsequently, the collected specimen is placed immediately in the same aliquot of the sterile viral transport medium (VTM).24,30 However, NP swab from both NP has become a preferred method of collecting the secretions since it is better tolerated and safer for the healthcare provider as they collect the secretions by standing beside the infected person. To collect the OP secretions, the healthcare provider has to be in front of the infected person to visualize the OP. Furthermore, it has been reported that specimens collected from the OP site showed a higher mean cycle threshold (CT) value than specimens obtained from the NP site suggesting a lower viral load in the OP.31 It has been stated that the OP site collection method is less sensitive than the NP site collection; however; more data is needed to confirm this observation.31
The healthcare provider collecting the specimens must be equipped with proper personal protective equipment (PPE) kit to prevent the risk of getting an infection. The NP swab must be inserted into the nasal cavity parallel to the hard palate between inferior turbinate and floor of the nasal cavity 1 inch past anterior nares till the patient flinches or resistance is encountered, indicating contact of the swab with the NP. The swab is kept in situ for 10 seconds to allow the secretions to adhere to the flocked tip. Subsequently, the swab is rubbed and rolled before being put into the VTM vial. Ideally, the swab should have flocked nontoxic synthetic fibers such as polyester and synthetic nylon handles.32 However, in an emergency, other nonflocked swabs and transport media may be used. Cotton swabs are usually avoided as they contain substances that may inactivate the virus and inhibit PCR reaction, thus affecting the test results. After collecting the sample, swabs should be placed immediately in labelled VTM vials; subsequently, these VTM vials should be packed triple-layered and transported directly to the laboratory in icepacks to maintain the temperature. After sample collection, proper doffing is done, whereas PPEs are disposed of properly into the yellow bag and sent for incineration as per guidelines.
The prompt transportation of the collected specimen in the VTM is desirable. Ideally, the specimen may be stored at 4°C for 24 to 72 hours or at −70 to −80°C for long-term storage, thus maintaining the cold chain.32 It is relevant to mention here that in some cases, specimens collected from saliva or the NP and OP sites may miss the early signs of infection, and also the late stage of infection. In lower respiratory tract infections and patients having acute respiratory distress syndrome, either repeated testing is done or sample is obttained from lower respiratory tract.
Identifying the patient's infection status depends on several factors, such as the site of the specimen, method of collection, the pathogen's burden at the collection site, and the severity of the disease. In addition, repeated sampling is done when the patient has clinical–radiological features of viral pneumonia with a history of potential exposure. Thus, it becomes difficult to interpret the result of a single negative test, especially among healthcare personnels.
Lower Respiratory Tract Infection of SARS-CoV-2
For lower respiratory tract infection, sputum or bronchoalveolar lavage should be collected as they yield the highest viral loads.33,34 Care must be taken while collecting these samples as the procedures are aerosol-generating, which may infect the healthcare personnel. Some patients with SARS-CoV-2 pneumonia have a high viral load in their fecal samples.35,36
Analytical Issues
Safety Issues for Specimen Processing for RT-PCR
As the virus is airborne, the processing should be done in class-2 Biosafety cabinets.24,25,37 However, it is always better to use a BSL-3 facility equipped with a negative pressure environment. The VTM containing respiratory viral specimens should be transferred to the lysis buffer in the BSL-2 cabinet. The lysis buffer contains a guanidinium-based inactivating agent and a nondenaturing detergent.38,39 Alternatively, when viral isolation is not required, the VTM may be added with guanidium salt. This process inactivates the viral particles and stabilizes the RNA. The specimen should not be exposed to 56°C as this temperature inactivates the SARS-CoV-2 and disrupts the RNA genome of the virus.
Many self-enclosed systems are integrating nucleic acid extraction, amplification and detection, such as ID NOW (Abbot, San Diego, California, United States), Cobas Liat (Roche Molecular System, Pleasanton, California, United States), and GeneXpert Dx (Cepheid, Sunnyvale, California, United States). Recently, on April 10, 2020, ICMR approved using TrueNAT for E gene screening for this novel SARS-CoV-2 infection. This E gene screening rules out all negative cases, whereas E gene-positive cases are further confirmed by the TrueNAT RdRp confirmatory test, which ICMR approves. This automatic system can easily verify the cases within 2 hours. This testing as a point of care is helpful in peripheral areas of developing countries where secretion collected from NP and middle inferior nasal turbinate can directly be placed in TrueNAT lysis buffer.
This equipment may be used as a point of care test after a clinical specimen gets transferred into a cartridge in a class-2 biosafety cabinet. The cartridge is sealed, and if any spill occurs, it should be decontaminated with an appropriate solution (usually 10% hypochlorite). However, in certain situations, especially when putting the testing system at the point of care, the biosafety cabinet of class-2 may not be available. In this case, the collection and transfer of specimen from a patient should be done by taking full precautions such as using the full protective gear (splash guard, N-95 mask, head cover, gloves, and disposable impervious laboratory coat).
Molecular Detection of SARS-CoV-2
The random amplification deep sequencing method is a particular way in the initial identification of SARS-CoV-2. Next-generation sequencing and metagenomic will be necessary to determine this virus's mutations.
Loop-mediated isothermal amplification, multiplexed isothermal amplification followed by microarray-based detection, and CRISPR (Clustered Interspersed Short Palindromic Repeats) assays may also be used in identification.40
Targeted real-time polymerase chain reaction
A real-time RT-PCR is recommended for molecular testings at a large scale as amplification and detection are done simultaneously in a closed system. RT-PCR avoids the possibility of PCR product contamination and allows real-time detection. Several coronaviruses are causing respiratory and intestinal infections in humans.2,41 Out of these coronaviruses, SARS-like bat corona, which includes both SARS-CoV-1 and SARS-CoV-2, is kept under one clade of the subgenus Sarbecoronaviridae.2,7 Of the several genetic markers, the structural genes are mostly targeted, for example, E, S, M helicase (Hel) and N, for detecting the SARS-CoV-2. In addition, some species-specific accessory genes, namely RNA-dependent RNA polymerase (RdRp) gene, ORF1a and ORF1b, HE, are targeted for confirmation. The WHO recommends using E gene for screening, whereas Centers for Disease Control and Prevention (CDC), United States, recommends the N1 and N2 genes. Due to the presence of other endemic coronaviruses and the potential genetic drift of SARS-CoV-2, two targets must be included for the testing. Therefore, mostly N, E, RdRp, and ORF1 genes are chosen for primer designing, and the results should be interpreted carefully.
Recently, the emergence of the Omicron VOC poses a great challenge for its diagnosis as there is an absence of the E gene. Therefore, we can miss the cases, caused by Omicron VOC. Other genes such as N, RdRp, and Orf1 can be targeted in such cases. Recently, CSIR-Central Drug Research Institute has developed an indigenous RT-PCR diagnostic kit, “Om,” to test the Omicron variant of coronavirus.42
Postanalytical Issues
Interpretation of Molecular Results
Usually, the current assays with three targets are employed to diagnose a case of COVID-19. To consider any sample positive for SARS-CoV-2, the positive peak for the E gene and with any one of the RdRp and ORF1 is deemed to be positive.43 However, CDC, United States, initially recommended if both the nucleocapsid targets N1 and N2 are positive, then the sample may be reported as positive for SARS-CoV-2.44 It is worth mentioning that the viral load does not reflect the severity of disease and also does not help in monitoring any therapeutic responses.26,27,28,45,46 Low CT values may be indicative of high viral loads and potentially more infections.33,47
Test for Cure and Infectivity
There is a suggestion that if two consecutive paired NP/OP samples collected 24 hours apart become negative by the RT-PCR detection method, the person may be considered free of the infection. Recently, one negative test result is also considered sufficient to discharge the admitted patients. Exceptions have been observed where many patients remain positive and shed viable/live coronavirus. Therefore, self-quarantine for 1 month may be recommended to help in containing the spread of SARS-CoV-2.19
During active multiplication in nasopharynx, virus can be isolated fromoropharynx and nasopharyns. However, despite having high viral load, virus may not isolated from their stool samples.29 Therefore, the present recommendation of ICMR is that the test for COVID-19 should be carried out on day 13 from the time of the isolation of the infected individual. The patient should be discharged from the isolation ward if the test is negative. The RT-PCR method speculates that SARS-CoV-1 could be detected for up to 1 month. A similar period may exist for SARS-CoV-2.2 Thus, it is highly recommended to have self-quarantined for up to 1 month after the discharge from the isolation ward.
Additional Methods of COVID-19 Testing
Serological Testing of COVID-19
The S and N proteins on the virus's outer surface come in direct contact with the immune system. Therefore, the serological method detects serum antibodies against S and N proteins.48 The spike protein is responsible for receptor binding and fusion. The fusion protein determines the tropism and transmission capabilities.2,7 The S gene coding this protein has two subunits, S1 and S2. The binding occurs through S1, while the fusion is executed through the S2 domain. SARS-CoV proteins bind to the angiotensin convertase enzyme-2 (ACE-2) receptor, but SARS-CoV-2 binds with greater affinity. This enzyme is abundant in human respiratory, renal, and gastrointestinal cells.2 The other protein is nucleocapsid, which plays an important role in pathogenesis, replication, and RNA packaging. N protein seems to be an immunodominant antigen as antibodies appear against N antigen.49
In about half of the symptomatic infected people, seroconversion usually occurs after 7 days, whereas approximately 97.8% of seroconversion happens after 21 days. However, SARS-CoV-2 viral load is observed in the stool samples, despite the appearance of serum antibodies.50 It has been noted that antibody detection is not associated with a decline in viral load. A rapid lateral flow assay determines serum (IgM and IgG) antibodies. The serological tests may be able to determine the immune status in the community against SARS-Cov-2. This host response-based detection is unlikely to be useful for early diagnosis.29,49 However, serology may be helpful as a surveillance tool for assessing herd immunity and confirming the community transmission of SARS-CoV-2 for surveillance purposes.36
Different antibody detection kits (lateral flow assay) have various sensitivity and specificity. Recently, ICMR-NIV Pune has developed an enzyme-linked immunosorbent assay (ELISA)-based IgG detection kit against SARS-CoV-2. In India, Zydus Cadila, Gujarat, is producing COVID KAVACH ELISA. However, ICMR has also approved this antibody-based detection kit only for surveillance among the high-risk population such as healthcare workers, frontline workers, or individuals belonging to the containment areas to identify the recovered infected persons.51
Antigen Detection
The rapid point of care immunoassay is generally lateral flow-based. These assays may detect antigens of the SARS-CoV-2 virus. Antigen detection would theoretically provide for the rapid and low-cost detection of SARS-CoV-2 but are likely to suffer from reduced sensitivity in early infection. The main concern with antigen detection is missing those cases with low viral load and variability in sampling. Recently, ICMR has approved using STANDARD Q COVID-19 Ag (antigen detection kit) to diagnose SARS-CoV-2 infection. The clinical samples having the SARS-CoV-2 antigen are to be considered infected, whereas RT-PCR should retest the negative results before declaring negative. For this, samples should be collected from the posterior pharynx of the suspected cases. The major advantage of this test is rapid as results are available within 30 minutes and can be executed at the point of care.
In 2021, ICMR approved the home testing of SARS-CoV-2 for only symptomatic individuals and immediate contacts of laboratory confirmed-positive cases. ICMR has reported these kits, CoviSelf (PathoCatch) COVID-19OTC Antigen LF device, PanBioCOVID-19 Antigen Rapid Test Device, CoviFindCOVID-19 Rapid Ag Self Test, AngcardCOVID-19 Home test kit, CliniTestCOVID-19 Antigen Self Test, ULTRA Covi-Catch SARS-CoV-2 Home Test, AbCheck Rapid Antigen Self Test – Nasal, COVID-19 AtHome Test Kit (Nasal), as satisfactory to be used as home/self test kits.50
Viral Isolation
This method is not recommended for diagnostic purposes since it requires a BSL-3 level facility. However, it has a role in researching and developing the diagnostic system, vaccines, and antiviral agents.
Disposal of the Generated Biomedical Waste
Different color-coded bins with foot-operated lids are to be kept where segregation and disposal of generated biomedical waste are done. The color-coded bins are labeled with COVID-19 waste. Used PPEs such as goggles, face-shield, single-use splash-proof apron, plastic coverall, and nitrile gloves are to be discarded into the red bag. In contrast, used masks (including triple layer masks, N95 masks), head cover/cap, shoe cover, and disposable linen gown are to be discarded into the yellow bags. VTM, plastic vials, vacutainers, Eppendorf tubes, plastic cryovials, and pipette tips are discarded into red bags.52
Conclusions
COVID-19 pandemic can be easily contained by early identification and isolation of the infected person. It has been observed that SARS-CoV-2 viral nucleic acid is usually absent in the serum and urine regardless of illness severity. However, the intact virus is maintained by respiratory specimens, sputum, saliva, NP swab, and OP swab collected in VTM. This can be detected by using the RT-PCR method. To date, this detection method is considered the most sensitive, reliable, and specific in detecting SARS-CoV-2 infection. However, the relevance of the serologic test is limited since it takes a more extended period to have detectable levels of antibodies in the blood samples. The other issue is cross-reactivity with related coronaviruses. However, antibody detection may be useful to determine the immunity of an individual and the community.
Similarly, antigen detection also traditionally suffers from suboptimal sensitivity. ICMR recommends testing three groups: hospitalized patients suspected of COVID-19, patients at high risk of poor outcomes, and persons who had close contact with someone with suspected or confirmed COVID-19. Testing in asymptomatic persons is not practical. Coinfection and secondary bacterial infection in COVID-19 have been reported. The other surrogate diagnostic parameters may be utilized, such as chest imaging by X-ray, computed tomography scan or magnetic resonance imaging, although they are not specific.
Acknowledgement
We gratefully acknowledge the support of Viral Research and Diagnostic Laboratory (State level- IMS BHU, Varanasi) which is funded by the Department of Health Research, Indian Council of Medical Research, India, in the preparation of this manuscript.
Conflict of Interest
None declared.
References
- The 2019 novel coronavirus disease (COVID-19) pandemic: a review of the current evidence. Indian J Med Res. 2020;151(2 & 3):147-159.
- [CrossRef] [PubMed] [Google Scholar]
- at: https://www.worldometers.info/coronavirus/country/india/ (accessed )
- Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23(08):468-478.
- [CrossRef] [PubMed] [Google Scholar]
- A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265-269. 10.1038/s41586-020-2008-3 [published correc tion appears in Nature. 2020; 580(7803):E7]
- [CrossRef] [PubMed] [Google Scholar]
- Molecular interactions in the assembly of coronaviruses. Adv Virus Res. 2005;64:165-230.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565-574.
- [Google Scholar]
- Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(06):e00473-e12.
- [CrossRef] [PubMed] [Google Scholar]
- Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(07):3995-4008.
- [CrossRef] [PubMed] [Google Scholar]
- The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120-133.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative genomic analyses reveal a specific mutation pattern between human coronavirus SARS-CoV-2 and Bat-CoV RaTG13. Front Microbiol. 2020;11:584717.
- [CrossRef] [PubMed] [Google Scholar]
- Overview of the pathogenesis of COVID-19 (Review) Exp Ther Med. 2021;22(03):1011.
- [CrossRef] [PubMed] [Google Scholar]
- SARS-CoV-2 pathogenesis. Nat Rev Microbiol. 2022;20(05):270-284.
- [CrossRef] [PubMed] [Google Scholar]
- Cardiac arrhythmias in patients with COVID-19. J Arrhythm. 2020;36(05):827-836.
- [CrossRef] [PubMed] [Google Scholar]
- COVID-19 infection: an overview on cytokine storm and related interventions. Virol J. 2022;19(01):92.
- [CrossRef] [PubMed] [Google Scholar]
- IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6(10):a016295.
- [CrossRef] [PubMed] [Google Scholar]
- Interleukin-8 production by macrophages from atheromatous plaques. Arterioscler Thromb Vasc Biol. 1996;16(08):1007-1012.
- [CrossRef] [PubMed] [Google Scholar]
- Transmission of SARS-CoV-2: implications for infection prevention precautions. at: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed )
- [Google Scholar]
- SARS-CoV-2 Variant Classifications and Definitions. at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed )
- [Google Scholar]
- Omicron has started replacing Delta variant in India, say sources. at: https://www.thehindu.com/news/national/omicron-has-started-replacing-delta-variant-in-india-say-sources/article38080589.ece (accessed )
- [Google Scholar]
- COVID-19—new insights on a rapidly changing epidemic. JAMA. 2020;323(14):1339-1340.
- [CrossRef] [PubMed] [Google Scholar]
- COVID-19—navigating the uncharted. N Engl J Med. 2020;382(13):1268-1269.
- [CrossRef] [PubMed] [Google Scholar]
- China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708-1720.
- [Google Scholar]
- Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(06):e00512-e00520.
- [CrossRef] [PubMed] [Google Scholar]
- Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10(02):311-316. 030759
- [CrossRef] [PubMed] [Google Scholar]
- Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(04):411-412.
- [CrossRef] [PubMed] [Google Scholar]
- SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177-1179.
- [CrossRef] [PubMed] [Google Scholar]
- Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(05):565-574.
- [Google Scholar]
- Author Correction: Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;588(7839):E35. DOI: 10.1038/s41586-020-2196-x
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. PLoS One. 2011;6(06):e21610. DOI: 10.1371/journal.pone.0021610
- [CrossRef] [PubMed] [Google Scholar]
- Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843-1844.
- [CrossRef] [Google Scholar]
- Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J Clin Microbiol. 2012;50(03):1064-1065.
- [CrossRef] [PubMed] [Google Scholar]
- Early transmission dynamics in Wuhan, China, of novel Coronavirus–infected pneumonia. N Engl J Med. 2020;382(13):1199-1207.
- [Google Scholar]
- Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71(15):793-798.
- [CrossRef] [PubMed] [Google Scholar]
- Singapore 2019 Novel Coronavirus Outbreak Research Team. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488-1494.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(01):386-389.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(04):549-555.
- [CrossRef] [PubMed] [Google Scholar]
- Virus inactivation by nucleic acid extraction reagents. J Virol Methods. 2004;119(02):195-198.
- [CrossRef] [PubMed] [Google Scholar]
- Inactivation and safety testing of Middle East respiratory syndrome coronavirus. J Virol Methods. 2015;223:13-18.
- [CrossRef] [PubMed] [Google Scholar]
- Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9(01):597-600.
- [CrossRef] [PubMed] [Google Scholar]
- Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9(01):747-756.
- [Google Scholar]
- CDRI develops cost-effective, indigenous Omicron testing kit. at: https://timesofindia.indiatimes.com/city/lucknow/cdri-develops-cost-effective-indigenous-omicron-testing-kit/articleshow/89078818.cms (accessed )
- [Google Scholar]
- New coronavirus pneumonia prevention and control protocol, 7th ed. National Commission of the People's Republic of China. (In Chinese.) 2020 at: http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf (accessed )
- [Google Scholar]
- Washington State 2019-nCoV Case Investigation Team. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929-936.
- [CrossRef] [PubMed] [Google Scholar]
- A well infant with coronavirus disease 2019 with high viral load. Clin Infect Dis. 2020;71(15):847-849.
- [CrossRef] [PubMed] [Google Scholar]
- Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(06):656-657.
- [CrossRef] [PubMed] [Google Scholar]
- Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect Control Hosp Epidemiol. 2020;41(05):493-498.
- [CrossRef] [PubMed] [Google Scholar]
- Secondary attack rate and superspreading events for SARS-CoV-2. Lancet. 2020;395(10227):e47. DOI: 10.1016/S0140-6736(20)30462-1
- [CrossRef] [PubMed] [Google Scholar]
- Advisory for COVID-19 Home Testing using Rapid Antigen Tests (RATs) at: https://www.icmr.gov.in/pdf/COVID/kits/COVID_Home_Test_Kit_13012022.pdf (accessed )
- [Google Scholar]
- Seroconversion among COVID-19 patients admitted in a dedicated COVID hospital: a longitudinal prospective study of 1000 patients. Med J Armed Forces India. 2021;77(Suppl. 02):S379-S384.
- [CrossRef] [PubMed] [Google Scholar]
- Guidelines for Handling, Treatment and Disposal of Waste Generated during Treatment/Diagnosis/Quarantine of COVID-19 Patients. at: https://cpcb.nic.in/uploads/Projects/Bio-Medical-Waste/BMW-GUIDELINES-COVID_1.pdf (accessed )
- [Google Scholar]






