Translate this page into:
Association of KCNJ11 gene (rs5219) polymorphism with HOMA-IR and HOMA B values in type 2 diabetes mellitus in India: A case-control study
*Corresponding author: Dr. Shilpa Suneja, Professor, Department of Biochemistry, Vardhman Mahavir Medical College and Safdarjung Hospital, New Delhi, India- 110029. Email: shilpasuneja@rediffmail.com
-
Received: ,
Accepted: ,
How to cite this article: Ramteke A, Suneja S, Muntakhab M, Gangopadhyay S, Kaur C. Association of KCNJ11 gene (rs5219) polymorphism with HOMA-IR & HOMA B values in type 2 diabetes mellitus in India: A case-control study. Ann Natl Acad Med Sci (India). 2024;60:218-24. doi: 10.25259/ANAMS-2023-4-15-(912)
Abstract
Objectives
Type 2 diabetes mellitus (T2DM) is a complex illness that results from either insulin resistance or insufficient insulin, which raises blood sugar levels. Numerous genes interact to influence the secretion of insulin. A gene of great interest is KCNJ11 of subfamily-J, member 11, which functions as an inwardly rectifying ATP-sensitive potassium (KATP) channel in pancreatic beta cells and is involved in glucose-stimulated insulin release.
Material and Methods
The present case-control study attempts to delineate the genetic impact of KCNJ11 (rs5219) gene polymorphism on the risk of T2DM in the Indian population. It involves 55 patients with type 2 diabetes (fasting plasma glucose of >126 mg/dl, 2-h glucose of >200 mg/dl, or HbA1c level of >6.4%) and 55 healthy controls (fasting plasma glucose of <100 mg/dl, 2-h glucose of <140 mg/dl, or HbA1c level of <6.4%). polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) was used to study KCNJ11 polymorphism through a standard protocol. Enzyme Linked Immunosorbent Assay (ELISA) was used to estimate serum Insulin levels. HOMA-IR & HOMA-β values were calculated. Statistical analysis was done using t-test, Chi-Square test, and One-way analysis of variance (ANOVA) test.
Results
Serum insulin levels and HOMA-IR values were significantly decreased in cases than in the control group. Logistic regression analysis showed that the frequency of KK genotype in T2DM individuals (21.8%) was higher than the control group (9%) (p = 0.01). Frequency of K allele (38%) in patients was higher than the control group (18%) (p = 0.001). The K allele risk in diabetic patients was 9.9 times higher as compared to controls (p = 0.001, OR 9.9, 95%Cl 0.036–0.36). Homeostatic model assessment β (HOMA-β) values of KK genotype (59.9±27.8315) were lower than that of EK (76.8±33.23) and EE (127.9±44.59) genotypes (p < 0.001).
Conclusion
The presence of KCNJ11 (rs 5219) gene polymorphism shows a noteworthy correlation with the likelihood of developing T2DM among the North Indian population. K allele is more likely to be present in individuals with T2DM than the control group. Moreover, HOMA-β values of those with the KK genotype were found to be lower than the individuals having EK and EE genotypes.
Keywords
KCNJ11
Type 2 Diabetes Mellitus
rs5219 gene polymorphism
Insulin
HOMA-β
HOMA-IR
INTRODUCTION
The multifactorial illness known as type 2 diabetes mellitus (T2DM) is characterized by either insufficient insulin secretion or insufficient insulin activity, which hinders the body’s ability to maintain glucose homeostasis. Worldwide, the incidence of T2DM is on the rise.1 With over 250 million cases worldwide, T2DM is more widespread than it was a few decades ago.2 T2DM etiology has been linked to interactions between many genetic determinants and environmental variables. A deeper knowledge of the genetic architecture of type 2 diabetes has been made possible by the identification of at least 75 distinct genetic loci for the disease over the course of the last ten years thanks to advancements in genetic association studies.3 Some genes that are particularly good indicators of T2DM susceptibility include those that encode proteins essential for pancreatic beta cell functions, such as those linked to insulin secretion. The route leading to the release of insulin begins with glucose inhibiting ATP-sensitive potassium (KATP) channels. This is followed by the β-cell membrane depolarizing, an increase in calcium ion inflow, and intracellular calcium stimulating the exocytosis of insulin-containing granules. This KATP channel is an inwardly-rectifying K+ channel composed of two structurally unrelated subunits of an octamer protein complex with four pores. Potassium inwardly-rectifying channel, subfamily J, member11 (KCNJ11) subunits are linked to four high-affinity sulfonylurea receptor (SUR1, ABCC8) subunits.4 Recent years have seen a great deal of attention in KCNJ11 as a potential gene for Type 2 diabetes. The NCBI (National Centre for Biotechnology Information) database for humans has 180 single nucleotide polymorphisms (SNPs) related to this gene. It spans two kilobases, is located at 11p15.1, contains one exon that encodes 390 amino acids, and controls the amount of insulin secreted by pancreatic beta cells in response to glucose.5 It has been demonstrated that the reduction in KATP sensitivity caused by the substitution of the amino acid lysine (K) for glutamate (E) in codon 23 of the KCNJ11 gene causes the channel to open for a longer period of time, hence reducing insulin production.6,7 As a result, several researches found a strong correlation between the E23K variation of KCNJ11 and T2DM susceptibility.8–10
Previous research reports have indicated that Asians seem to have a fairly common KCNJ11 polymorphism (rs5219). Small and medium-sized researches were conducted in this community, and conflicting findings about the link between this variation and type 2 diabetes were found.11–16 The influence of rs5219 on susceptibility to type 2 diabetes is not fully explored due to the published research’s limitations in terms of sample size and ethnic diversity, and their small sample size may make it impossible to draw meaningful conclusions. The conclusive determination of the connection between KCNJ11 polymorphism (rs5219) and T2DM in South Asian populations requires the completion of sufficient thorough meta-analyses and genome-wide association studies (GWAS).
The current study was conducted to assess the relationship between KCNJ11(rs5219) gene polymorphism and associations between this genetic variation and diabetes susceptibility in T2DM in India, as the genetic background for this genetic variant has not been investigated much for T2DM in this population.
MATERIAL AND METHODS
Study Population
This case-control study was done in the Department of Biochemistry and the Department of Medicine, V.M.M.C. & Safdarjung Hospital in New Delhi, India, after clearance from the Ethical Committee of Vardhman Mahavir Medical College and Safdarjung Hospital. Informed written consent was obtained from each subject before enrolling them in the study. The sample size was calculated using the formula as shown below.17
Calculation
Where n is the sample size.
PA is proportion of cases and PB is proportion of controls.
According to the above formula, the sample size is calculated as 113 by taking the finite population of T2DM to be 200. In view of hospital statistical data and after inclusion and exclusion criteria, the minimum sample size is kept as 55 cases and 55 controls.
The study subjects included 55 patients having T2DM and 55 healthy age- and sex-matched controls. The diagnosis of type 2 diabetes was done on the basis of symptoms of polyuria, polydipsia, and polyphagia. Relevant investigations included fasting plasma glucose more than 126 mg/dl, plasma glucose more than 200 mg/dl after 2-h oral glucose (1.75 g/kg), or HbA1c ≥ 6.5%.18
Exclusion criteria
Patients with type 1 diabetes mellitus or gestational diabetes, pancreatitis, patients on oral hypoglycaemic and insulin therapy, patients with other endocrine diseases and impaired renal and hepatic function, and those receiving hypercholesterolemia, hypertension, or corticosteroid medications were excluded from the study.
Sample collection
After an overnight fast, study participants had their blood drawn via venepuncture. Each of the three vacutainer types—Ethylenediamine tetraacetic acid (EDTA), plain (without an anticoagulant), and NaF—was used for the collection of 2 mL of blood. Samples of EDTA were kept at −20°C to extract DNA. After allowing the blood in the ordinary vacutainer to coagulate, the serum was extracted and centrifuged for additional examination. Prior to batch analysis for the enzyme assay, these samples were kept at −70°C. As soon as the blood was drawn, it was utilized to estimate serum glucose in a NaF vacutainer.
Biochemical measurements
Biochemical parameters, fasting blood Glucose, and postprandial glucose levels were processed using Siemens ADVIA Germany, a fully automated chemistry analyzer.
Estimation of Serum Insulin Levels, HOMA-IR, and HOMA-β
Serum insulin levels were estimated using a commercially available enzyme-linked immunosorbent assay (ELISA) kit manufactured by Calbiotech Inc (Catlog no. P1099D), following the instructions from the manufacturer. This utilizes a two-step protocol for enzyme immunoassay in quantitative measurement of serum insulin. The absorbance was read at the wavelength of 450 nm. The optical density that was measured was directly proportional to the insulin concentration. Homeostasis Model Assessment IR (HOMA-IR) index is calculated by the product of basal glucose and insulin levels divided by 22.5,19 and is considered to be a simple, inexpensive, and reliable surrogate measure of insulin resistance. Homeostasis Model Assessment β (HOMA-β) value was determined as shown below in the formula to analyze fasting insulin level reflecting pancreatic β-cell function.
Genotyping
DNA Extraction Kit (Krishgen) was used to extract genomic DNA from nucleated blood cells. The extracted DNA was amplified using polymerase chain reaction (PCR) by utilizing the following primers- forward=5’‐GACTCTGCAGTGAGGCCCTA‐3’ reverse=5’ACGTTGCAGTTGCCTTTCTT‐3’.20 The Indian company “Eurofins Scientific” created the primers. The amplification of DNA was done using a PCR reaction mixture with the help of a thermal cycler (Himedia Eco-96). This 25 µL PCR reaction mixture contained 3 µl of genomic DNA, 0.25 µl of each primer, 6.5 µl of nuclease-free water, and 10 µL of Dream Taq green PCR Master-mix manufactured by Thermo fisher-scientifc (Catalog no. K1081), which had 3.2 µM Taq DNA polymerase, 3.2 µM 2X bufer, 32 µM MgCl2, and 3.2 µM of each dNTPs. The following PCR conditions were used: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 sec; annealing at 57°C for 30 sec, then extension at 72°C for 30 sec. The final extension step was at 72°C for 5 min and kept for temporary storage at 4°C. The PCR amplification was confirmed by gel electrophoresis (2% agarose gel and Ethidium bromide staining). The amplified PCR product of 210 bp in 1 μl quantity was incubated with BanII (Catlog no. R0119S) (New England, Biolabs, England) restriction enzyme20 at 37°C for 60 min in 50 μl reaction mixture that contained 10 × NE buffer. The digested PCR products were then resolved on 2% agarose gel electrophoresis for the E23K genotypes. The Homozygous KK Genotype was digested into two fragments of 178 bp and 28 bp. Homozygous EE Genotype was cut into three fragments of 150 bp, 32 bp, and 28 bp in heterozygous subjects. With the EK genotype, the PCR product breaks into four fragments of 178 bp, 150 bp, 32 bp and 28 bp. On 2% agarose gel electrophoresis, 32 bp and 28 bp being very small are not visible on gel as depicted in Figure. 1.
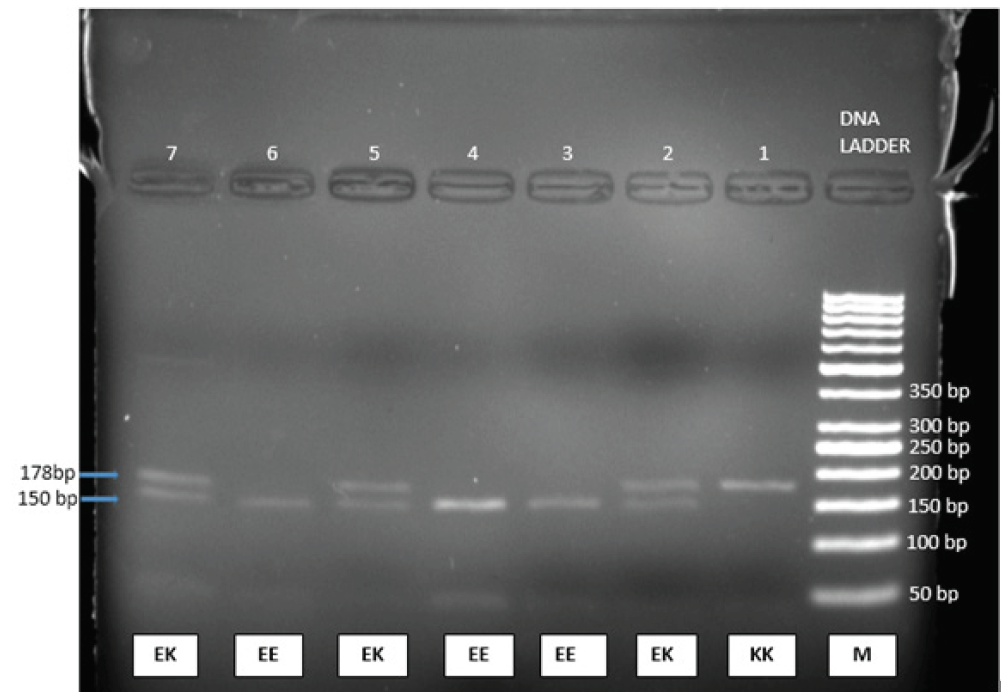
- KCNJ11 gene polymerase chain reaction (PCR) products were broken down using the BanII restriction enzyme. At 150 bp, homozygous EE has a single band. There are two bands at 178 bp and 150 bp in heterozygous EK. The homozygous KK sample was electrophoresed on a 2% agarose gel, revealing a single band at 170 bp. Lane 1 displays the homozygote KK genotype, lanes 2, 4, and 7 the heterozygote EK genotype, and lanes 3, 4, and 6 the homozygote EE genotype. M is a DNA ladder (50 bp ladder). Due to their extremely small size, the bands corresponding to sizes 32 and 28 bp are invisible in the gel.
Statistical analysis
The Statistical Package for the Social Sciences for Windows (SPSS), version 25.0 (Chicago, IL, USA), was used to do the final analysis after the data were imported into a Microsoft Excel spreadsheet. The categorical variables were displayed as percentages (%) and numbers. The means ± SD were used to present the quantitative data. P values less than 0.05 were regarded as statistically significant. Using the Chi-Square test, any divergence from the Hardy–Weinberg equilibrium was assessed. A 95% confidence interval (CI) was used to compute the odds ratio (OR) in order to assess the relationship between T2DM genotypes and the KCJN11 gene polymorphism. The Independent t-test was utilized to assess quantitative variables. The Chi-Square test was used to assess qualitative characteristics.
Results
This study confirmed the association between KCNJ11 (rs5219) gene polymorphism in HOMA-IR and HOMA-β values in 110 participants. Of these, 55 patients were diagnosed with T2DM. As per ADA criteria, mean age was 46 ± 10.24 years where 40% were males and 60% were females. The control group comprised 55 age- and gender-matched healthy individuals with mean age of 42 ± 10.7 years, where 47.27% were males and 52.73% were females. The demographic profile along with the clinical and laboratory parameters of the study population is depicted in Table 1. The difference in age and gender between cases and controls was not statistically significant. However, weight and body mass index were significantly higher in cases than controls (p<0.001). Subgroup analysis of biochemical markers between T2DM and control group in this study showed statistically significant difference in HbA1C (p<0.001), fasting blood glucose (p<0.01), and postprandial blood glucose (p<0.001) with values greater than in the control group.
| Characteristics | Cases (n=55) | Controls (n=55) | P-value |
|---|---|---|---|
| Age (years) | 46±10.24 | 42.34±10.7 | 0.071 |
| Males, % | 40 | 47.27 | 0.442 |
| Weight (kg) | 72.65±10.76 | 62.15±8.58 | <.0001 |
| Height (m) | 1.6±0.04 | 1.8±0.02 | 0.678 |
| BMI (kg/m2) | 28.98±3.87 | 24.75±3.45 | <0.0001 |
| HbA1C (%) | 7.8±0.57 | 4.3±0.2 | <0.0001 |
| FBS (mg/dL) | 104.9±13.2 | 90.3636±9.6 | 0.01 |
| PP (mg/dL) | 189±13.8 | 127±2.4 | <0.0001 |
Data are expressed as Mean ± SD or percentage.
HbA1C: Glycated hemoglobin, BMI: Body mass index, FBS: Fasting blood sugar, PP: Postprandial blood sugar
Table 2 depicts the serum insulin levels, HOMA-IR, and HOMA-β values in T2DM patients and controls. Serum insulin levels were found to be significantly lower in cases than in the control group (9.95±3.12 vs 15.1±4.6 µIU/mL, p<0.0028). Similarly, HOMA-IR and HOMA-β and values showed a significant decrease in cases than controls (2.6±0.9 vs 3.3855±1.1 µIU/mL, p <0.03) and (2.6±0.9 vs 3.3855±1.1 µIU/mL, p <0.03), respectively.
| Study Group | Insulin (µIU/mL) | HOMA-IR (%) | HOMA-β (%) |
|---|---|---|---|
| T2DM | 9.95±3.12 | 2.6±0.9 | 96.3255±47.7 |
| Control | 15.1±4.6 | 3.3855±1.1 | 224.56±107.5 |
| P value | <0.0028 | <0.03 | <0.0001 |
T2DM: Type 2 diabetes mellitus, HOMA-IR homeostasis model assessment IR.
HOMA-β homeostasis model assessment β.
Table 3 displays the distribution of genotypes and alleles for the KCNJ11 rs 5219 (E23K) polymorphism. In the T2DM group, the frequencies of the EE, EK, and KK genotypes were 25%, 18%, and 12%, respectively, compared to 40%, 10%, and 5% in the control group. The homozygous KK genotype of KCNJ11 rs 5912 SNP has a 3.8-fold greater risk of type 2 diabetes in the Indian population, according to logistic regression analysis (odds ratio OR = 3.84, 95% Cl 1.21–12.21, p = 0.01). In the Indian population, the heterozygous EK genotype likewise demonstrated a 2.8-fold elevated risk of type 2 diabetes (odds ratio OR = 2.88, 95% Cl 1.15–07.22, p = 0.02). Individuals with type 2 diabetes showed a noticeably higher prevalence of the K allele than in controls (OR 9.9, 95% Cl 0.036–0.36. p = 0.001).
| Cases | Controls | P-value | OR | (95% CI) | |
|---|---|---|---|---|---|
| Genotypic frequency | |||||
| EE | 25 (45.45%) | 40 (80%) | Reference | ||
| EK | 18 (32.73%) | 10 (18%) | 0.02 | 2.88 | 1.15–7.23 |
| KK | 12 (21.82%) | 5 (9%) | 0.01 | 3.84 | 1.21–12.21 |
| Total | 55 | 55 | |||
| Genotypic frequency | |||||
| EE | 25 (45.45%) | 40 (72%) | 7.37 (0.06–0.47) | ||
| EK & KK | 30 (55%) | 15 (27%) | 0.0066 | 2.88 | |
| Total | 55 | 55 | |||
| Allelic frequency | |||||
| E | 68 (62%) | 90 (82%) | 0.001 | 9.9 | 0.036–0.36 |
| K | 42 (38%) | 20 (18%) | |||
OR: Odds ratio, Cl: Confidence interval.
The inter-genotypic variation of the HOMA-β in T2DM and controls is shown in Table 4. Our results showed that HOMA-β in KK, EK, and EE genotypes of the patients with T2DM was 59.9±27.83, 76.8±33.23, 127.9±44.60%, respectively. The inter-genotypic difference in HOMA-β values was statistically significant in T2DM patients.
| Variable | Mean ± SD | P | ||
|---|---|---|---|---|
| KK (n=12) | EK (n=18) | EE(n= 25) | ||
| HOMA-β (%) | 59.9± 27.8315 | 76.8± 33.2314 | 127.9± 44.5975 | <0.0001 |
Values are expressed as Mean ± standard deviation (SD)
DISCUSSION
One of the most common noninfectious diseases in the world, type 2 diabetes has been getting more and more common over the past few decades. Susceptibility to T2DM is influenced by both environmental susceptibility and genetic background. The illness has been linked to several genes.21 The KCNJ11 protein encodes a crucial role in the production of insulin from pancreatic β-cells, suggesting that it may be a susceptibility gene for type 2 diabetes. However, E23K polymorphism in KCNJ11, plays an important role in suppression of insulin secretion.22,23 The amino acid substitution, replacing lysine (AAG) with glutamic acid (GAG) resulting due to E23K polymorphism leads to altered physiochemical properties wherein the negative charge replace the positive charge. This prolongs the opening of KATP channel and decreases its sensitivity to ATP and suppresses insulin production.24,25
When stratified by ethnicity, a meta-analysis study by Takeuchi et al.26 revealed a significant connection between the E23K (rs5219) polymorphism and T2D risk among East Asians and Caucasians; no significant associations were detected among South Asians and other ethnic populations. Given the variability in associations and scarcity of literature from the Indian subcontinent, we tried to investigate the correlation of this SNP with HOMA-β and HOMA-IR in the Indian population for T2DM patients and controls.
Our findings support previous research on the Russian, Syrian, and Iranian populations, which indicated that patients had a higher frequency of the KK genotype than controls.27–29 In the study done by Li YY in 2013,29 it was found that E23K contributed to T2DM susceptibility.
In addition, our data showed that KK genotype carriers were significantly more likely than controls to develop type 2 diabetes. Furthermore, HOMA-β was used as a model to analyze pancreatic β-cell activity and evaluate the insulin secretion of the study participants. The results showed that there was a statistically significant difference in HOMA-β values between the KK, EK, and EE genotype groups. These findings are consistent with the research conducted by Sunita et al.30 Our study’s K allele prevalence of 38% is comparable to that of research conducted in Asian and Caucasian populations.31–34
A range of models were used to assess the relationship between the KCNJ11 rs5219 polymorphism and the risk of type 2 diabetes. The results showed that the dominant genetic model (EE vs. KK + EK) was linked to a sevenfold increased risk of developing diabetes compared to controls, with an OR of 7.37 (95% CI 0.06 to 0.47, P = 0.006). Wang et al.’s meta-analysis study34 indicates that the KK genotype is consequently linked to an eightfold increase in the risk of T2DM. In a similar vein, other studies have also been published30,35–38 showing a link between the KK genotype and an increased risk of diabetes.
The KK genotype’s HOMA-β values (59.9±27.83) in our study were found to be lower than those of the EK (76.8±33.23) and EE (127.9±44.60) genotypes. A One-Way ANOVA analysis revealed a significant difference (P<0.0001) in the HOMA-β value between the EE, EK, and KK genotypes. The effects of a single genetic component on the development of T2DM varies throughout individuals due to differences in environmental circumstances. T2DM is recognized as a complex condition caused by the interaction of several hereditary and environmental factors. In India, a large number of people experience varying degrees of stress and generally have poor eating habits. This might account for the high-risk value (nine times) in our study in a sample of the Indian population in comparison to other groups, since these potent environmental factors may magnify the influence of the genetic component in the risk allele carriers.
CONCLUSION
This study showed an association between rs5219 polymorphism of the KCNJ11 gene with susceptibility to develop T2DM in the Indian population. Also, K allele is more likely to be present in individuals with T2DM than the control group. However, additional studies with a larger sample size will be required to confirm this marker in the Indian population.
The limitation of the present study is that it did not consider the interactions between the gene and its protein. The study was conducted only on a single SNP of the KCNJ11(rs5219) gene. Hence, further studies on other SNPs of KCNJ11 should be carried out to prove the significant association between genetic markers and biochemical markers in T2DM. Moreover, a large cohort study is required to validate the expression of these genes in diverse populations in a country like India.
Ethical approval
The research/study is approved by the Institutional Ethics Committee at Vardhman Mahavir Medical College and Safdarjung Hospital, number IEC/VMMC/SJH/Project/2021-10/CC-192, dated 06th November 2021.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation:
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocr Rev. 1999;20:101-35.
- [CrossRef] [PubMed] [Google Scholar]
- Pharmacogenetics of anti-diabetes drugs. Pharmaceuticals. 2010;3:2610-46.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med. 2016;48:220.
- [Google Scholar]
- Toward understanding the assembly and structure of KATP channels. Physiol Rev. 1998;78:227-45.
- [CrossRef] [PubMed] [Google Scholar]
- Association of KCNJ11 (E23K) gene polymorphism with susceptibility to type 2 diabetes in Iranian patients. Adv Biomed Res. 2015;4:1-5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Association of KCNJ11 and ABCC8 genetic polymorphisms with response to repaglinide in Chinese diabetic patients. Acta Pharmacol Sin. 2008;29:983-9.
- [CrossRef] [PubMed] [Google Scholar]
- Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia. 2005;48:2544-51.
- [CrossRef] [PubMed] [Google Scholar]
- KIR6. 2 polymorphism predisposes to type 2 diabetes by inducing overactivity of pancreatic β-cell ATP-sensitive K+ channels. Diabetes. 2002;51:875-9.
- [CrossRef] [PubMed] [Google Scholar]
- Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the diabetes prevention program. Diabetes. 2007;56:531-6.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Genomic variation in pancreatic ion channel genes in Japanese type 2 diabetic patients. Diabetes Metab Res Rev. 2001;17:213-6.
- [CrossRef] [PubMed] [Google Scholar]
- Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53:1360-8.
- [CrossRef] [PubMed] [Google Scholar]
- Sequence variations in the human Kir6. 2 gene, a subunit of the beta-cell ATP-sensitive K-channel: no association with NIDDM in white Caucasian subjects or evidence of abnormal function when expressed in vitro. Diabetologia. 1996;39:1233-6.
- [CrossRef] [PubMed] [Google Scholar]
- Amino acid polymorphisms in the ATP-regulatable inward rectifier Kir6. 2 and their relationships to glucose-and tolbutamide-induced insulin secretion, the insulin sensitivity index, and NIDDM. Diabetes. 1997;46:508-12.
- [CrossRef] [PubMed] [Google Scholar]
- Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS biology. 2003;1:e20.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Association studies of variants in the genes involved in pancreatic β-cell function in type 2 diabetes in Japanese subjects. Diabetes. 2006;55:2379-86.
- [CrossRef] [PubMed] [Google Scholar]
- The effect of KCNJ11 polymorphism on the risk of type 2 diabetes: a global meta-analysis based on 49 case-control studies. DNA and cell biology. 2012;31:801-10.
- [CrossRef] [PubMed] [Google Scholar]
- Association of KCNJ11 rs5219 gene polymorphism with type 2 diabetes mellitus in a population of Syria: a case-control study. BMC Med Genet. 2019;20:1-6.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Standards of Medical Care in Diabetes 2011. Diabetes Care. 2011;34:S11.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Study of beta-cell function (by HOMA model) in metabolic syndrome. Indian J Endocrinol Metab. 2011;15:S44-S49.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Polymorphism E23K (rs5219) in the KCNJ11 gene in Euro-Brazilian subjects with type 1 and 2 diabetes. Genet Mol Res. 2017;16:1-9.
- [Google Scholar]
- Dong: Polymorphism rs189037C> T in the promoter region of the ATM gene may associate with reduced risk of T2DM in older adults in China: A case control study. BMC Med Genet. 2017;18:84.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- KCNJ11: Genetic polymorphisms and risk of diabetes mellitus. J Diabetes Res. 2015;2015:908152.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Molecular cell biology of KATP channels: implications for neonatal diabetes. Expert Rev Mol Med. 2007;9:1-17.
- [CrossRef] [PubMed] [Google Scholar]
- B-cell hyperexcitability: From hyperinsulinism to diabetes. Diabetes Obes Metab. 2007;9:81-8.
- [CrossRef] [PubMed] [Google Scholar]
- Independent case-control study in KCNJ11 gene polymorphism with Type 2 diabetes. Mellitus. 2022;29:2794-9.
- [Google Scholar]
- Ethnic difference in patients with type 2 diabetes mellitusin inter-East Asian populations: A systematic review andmeta-analysis focusing on gene polymorphism. Journal of Diabetes.. 2009;1:255-62.
- [CrossRef] [PubMed] [Google Scholar]
- The KCNJ11 E23K and ABCC8 exon 31 variants contribute to susceptibility to type 2 diabetes, glucose intolerance and altered insulin secretion in a Russian population. Diabetes Metab Syndr Clin Res Rev. 2008;2:185-91.
- [Google Scholar]
- Association of KCNJ11 (E23K) gene polymorphism with susceptibility to type 2 diabetes in Iranian patients. Adv Biomed Res. 2015;4:1.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The KCNJ11 E23K gene polymorphism and type 2 diabetes mellitusin the Chinese Han population: A meta-analysis of 6,109 subjects. Mol BiolRep. 2013;40:141-6.
- [Google Scholar]
- Lower HOMA-β values are detected among individuals with variant of E23K polymorphism of potassium inwardly-rectifying channel, subfamily J, member 11 (KCNJ11) gene. Egyptian J. Med Human Genetics. 2015;16:227-31.
- [Google Scholar]
- Classification and diagnosis of diabetes: Standards of medical care in diabetes-2018. Diabetes Care. 2018;41:S13-S27.
- [CrossRef] [PubMed] [Google Scholar]
- Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with type 2 diabetes mellitus (UKPDS 53) Diabet Med. 2001;18:206-12.
- [CrossRef] [PubMed] [Google Scholar]
- The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52:573-7.
- [CrossRef] [PubMed] [Google Scholar]
- Association of Kir6.2 gene rs5219 variation with type 2 diabetes: A meta-analysis of 21,464 individuals. Prim Care Diabetes. 2018;12:345-53.
- [CrossRef] [PubMed] [Google Scholar]
- The KCNJ11 E23K and ABCC8 exon 31 variants contribute to susceptibility to type 2 diabetes, glucose intolerance and altered insulin secretion in a Russian population. Diabetes Metab Syndr Clin Res Rev. 2008;2:185-91.
- [Google Scholar]
- KCNJ11 E23K affects diabetes risk and is associated with the disposition index: Results of two independent german cohorts. Diabetes Care. 2008;31:87-9.
- [CrossRef] [PubMed] [Google Scholar]
- Association of E23K (rs5219) polymorphism in the KCNJ11 gene with type 2 diabetes mellitus risk in Jordanian population. Human Gene.. 2023;37:201201.
- [Google Scholar]
- The KCNJ11-E23K gene variant hastens diabetes progression by impairing glucose-induced insulin secretion. Diabetes. 2021;70:1145-1156.
- [CrossRef] [PubMed] [Google Scholar]






